Hi, I’m using SDF reader (configure to Extract Mol blocks - use molecule name as row ID, extract molecule name) and passing the SDF file to Molecule to CDK module. On the console I get Format conversion failed afetr executing…
Any suggestions how to handle this error.
Thanks,
RG
Can you post the sd file you’re trying to manipulate?
2 Likes
THanks again…
TEST.txt (11.6 KB)
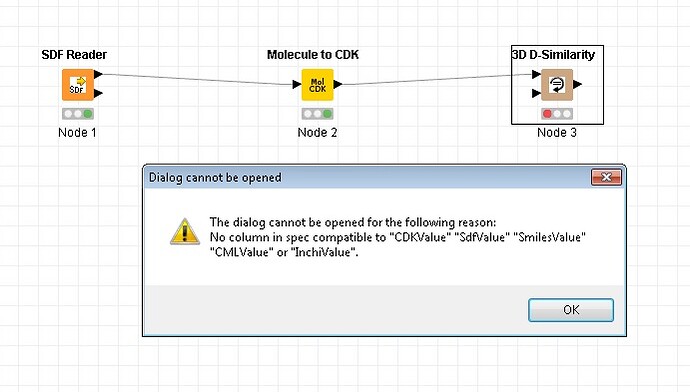
So, I read the sdf file. Molecule to CDK runs the samples fine (but on the console I see format conversion failed). When I try to go from Molecule to CDK to 3D D-Similarity and try to configure I get the error message on the picture attached.
THanks,
RG
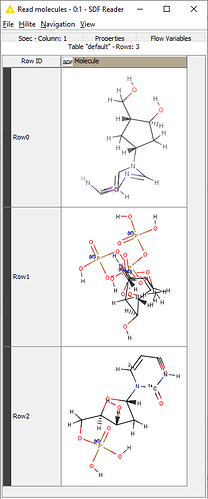
Hi @rolan1014, The structures in the file you uploaded are all quite messy. Where did you get them from?
On my machine the Molecule to CDK node works fine, but I wouldn’t trust those structures for any kind of structural analysis.
As for your new error message, the 3D D-Similarity node requires 2 inputs. You have only specified one. You need to add a second table containing the reference structure(s) before the node can be configured.
1 Like
Thanks for your reply Samuel…I will try that.
I got the list from the CAS registry. I applied RDKit optimize geometry, structure normalizer and added H to the initial list…Re-evaluate the initial work-flow?
THanks again…
Rolan