Also, @nemad - can you please elaborate on the "Your images have
to be normalized between [0, 1] " - I have 2K+ images; is this performed by hand? Any detail you can provide is appreciated, thanks.
Here is an updated version of the workflow that includes the normalization as well as some additional preprocessing for deep learning:
TransferLearning.knwf (38.2 KB)
Best,
nemad
Hi @nemad - I finally integrated python to Knime.
I was trying to initialize the DL Python Network Creator, but I get this error:
ImportError: No module named ‘keras’
ERROR DL Python Network Creator 0:2 Execute failed: Traceback (most recent call last):
File “C:\Program Files\KNIME\plugins\org.knime.python2_3.6.2.v201811051558\py\PythonKernelBase.py”, line 278, in execute
exec(source_code, self._exec_env, self._exec_env)
File “”, line 2, in
ImportError: No module named ‘keras’
I believe I installed Keras over the weekend; forgot how to check in cmd.
@asrichardson please check knime.com/deeplearning for instructions. Keras has to be installed in version 2.1.6 in your py35_knime environment.
Hi @nemad. I made some great progress - I was able to fully establish my Python and Keras environment per the help of @christian.dietz.
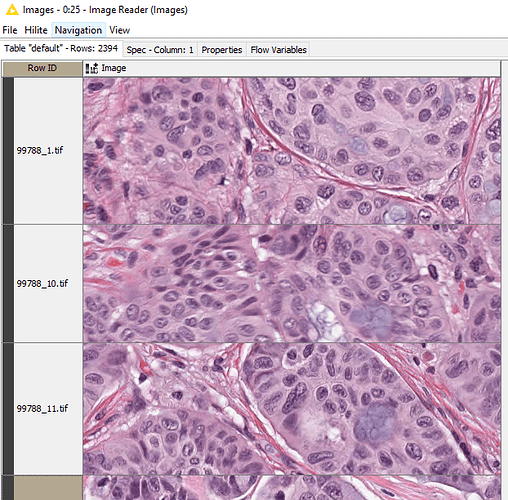
First: I’m having some issues with how I “read” and label my images via the “joiner”. I want to create a table that has (4) columns: Image Name, Image, RID, Score. Currently, I have (1) table and a set of images I want to join, they are in screenshots below. My join is adding “RowID” after the image name, today, which is not what I want.
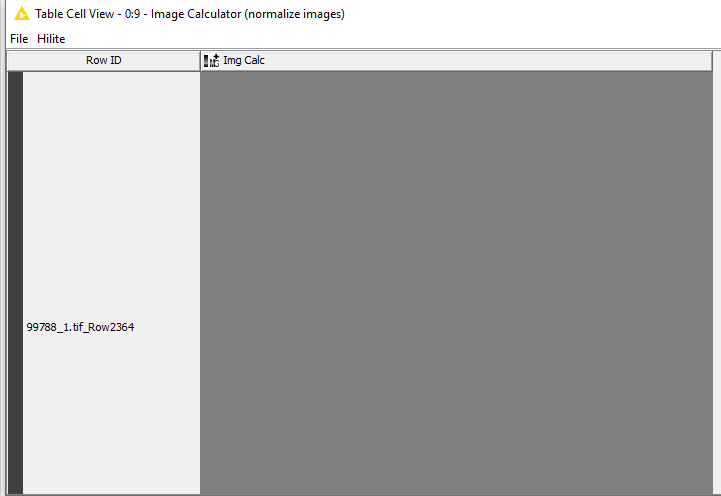
Second: The “image calc” node turned the images 100% grey - they are all the same. Not sure of the function that needs to happen here.
Third : I was able to convert RID to String, but I’m at a loss for how to manipulate this in the Domain Calculator node.
I’ve attached my workflow for reference.
Keras_Transfer_Learning.knwf (32.0 KB)
Any thoughts are appreciated, thanks.
-Anthony
Hello Anthony,
I am not sure if I understand your first problem.
You can simply use the joiner node to join the image table with the table containing the labels by specifying that the joining column for your image table is the Row ID and the joining column for your labels table is the ImageValues column.
The images are now converted to float values within the range [-1, 1], unfortunately it is currently not possible to see the actual content in the KNIME table view because it only supports values in [0, 255] for display. However, if you open the view of the Image Calculator and click on an image you will see that the images are still valid.
What you need to do for the rid column is to add the possible values to its domain.
You do this by making sure that it is in the include list and the checkbox “Restrict number of possible values” is NOT checked. Then you can use the one-to-many node to create the “one-hot” columns for the rid column that will serve as target for your classification.
Let me know if this helps 
Best,
nemad
Thanks @nemad
I’m getting close - I was able to join correctly, perform the image calc, and execute the domain calc w/ one-to-many hotshot.
My issue is now with the Keras Network Learner; I get this issue:
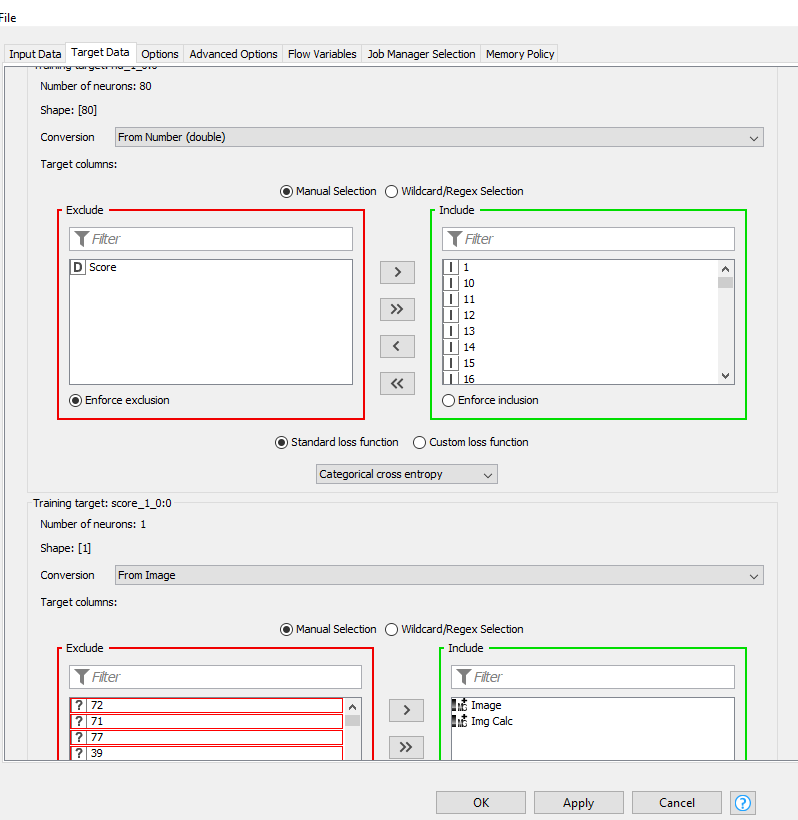
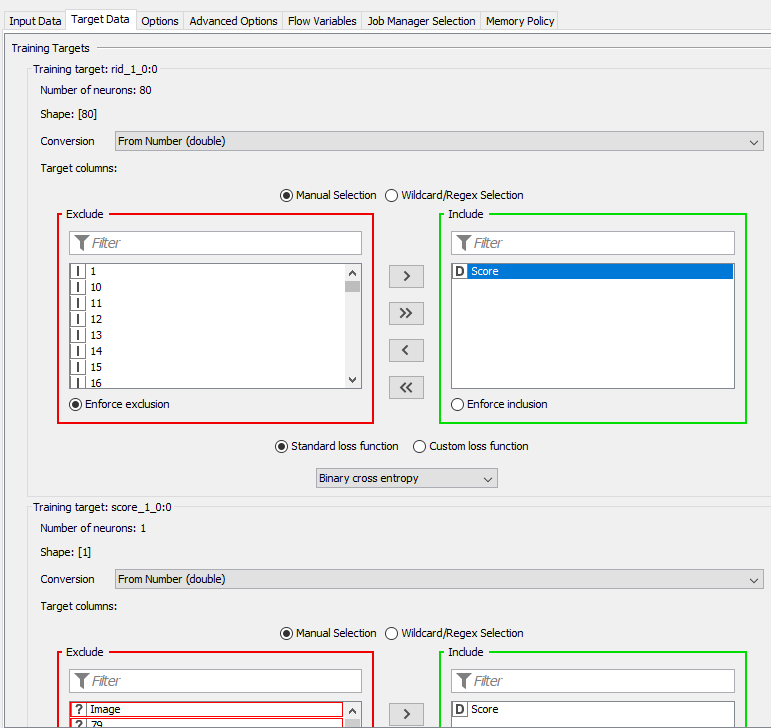
WARN Keras Network Learner 3:6 failed to apply settings: More target columns selected (2) than neurons available (1) for network target ‘score_1_0:0’. Try removing some columns from the selection.
This is my configuration:
As I get deeper into this, I’m starting to realize that maybe it would be better if I split the problem into (2); first, tackle RID, and then tackle score after, then just document the predictions in excel after (copy & paste).
This is a hard problem from my advisor, so it is not supposed to be easy 
Thanks,
Anthony
Hello Anthony,
your inputs for the images and the score target are incorrect.
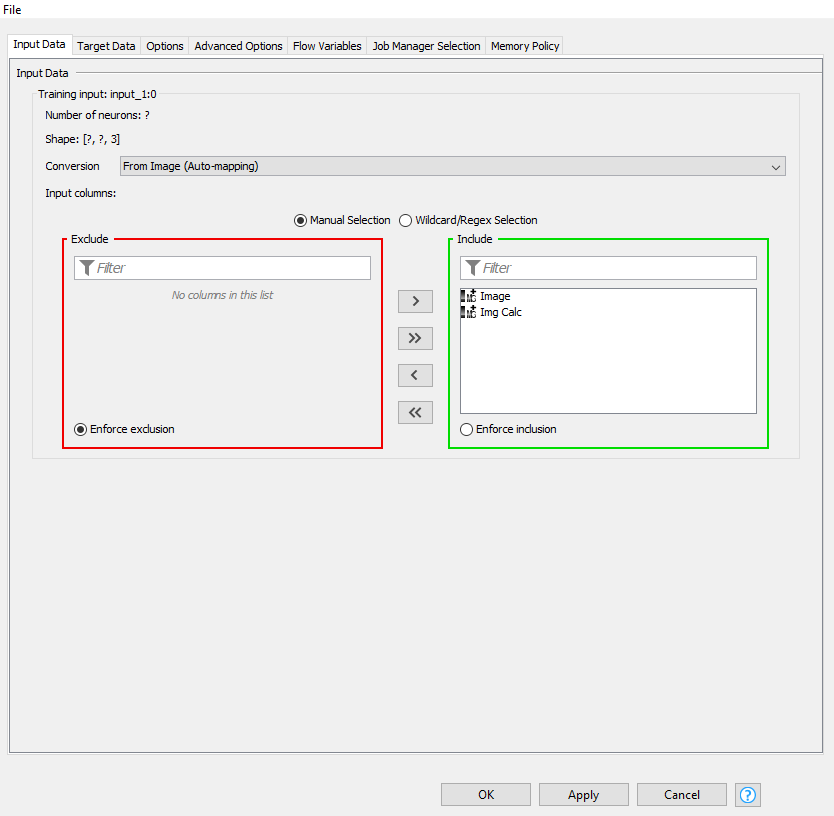
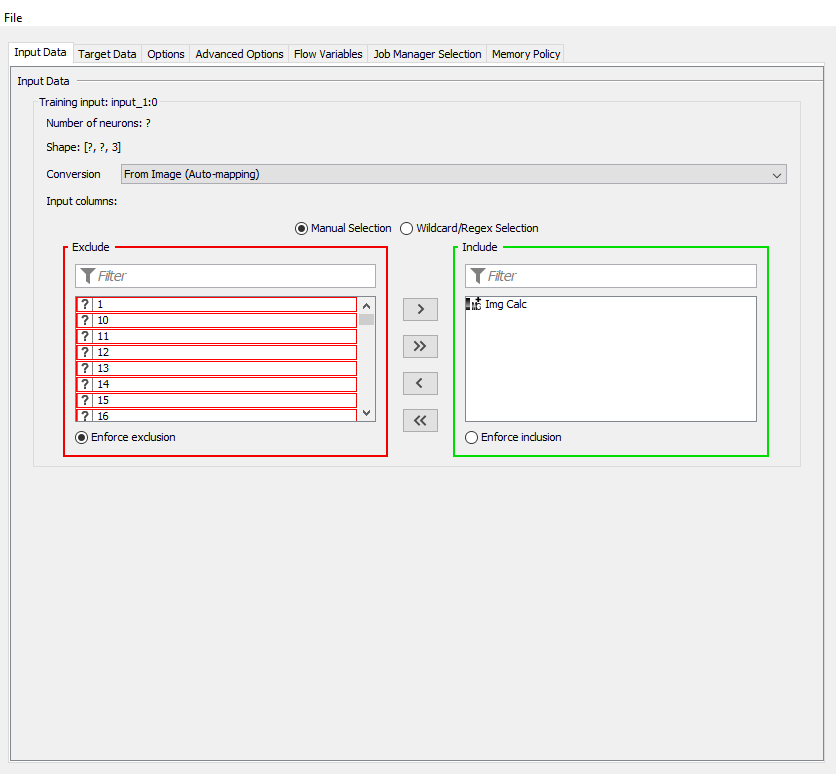
For the image, you need to only include the Img Calc column and for the score, you will have to switch the converter to “From Number (double)”, you should then be able to include the score variable.
Make sure that you select “Binary Cross Entropy” as the loss function for the Score target.
Don’t give up now, you are almost there 
That is not to say that you shouldn’t also try different strategies but I would suggest to first get this approach to work, as you already put a lot of effort into it.
Best,
nemad
Hi @nemad,
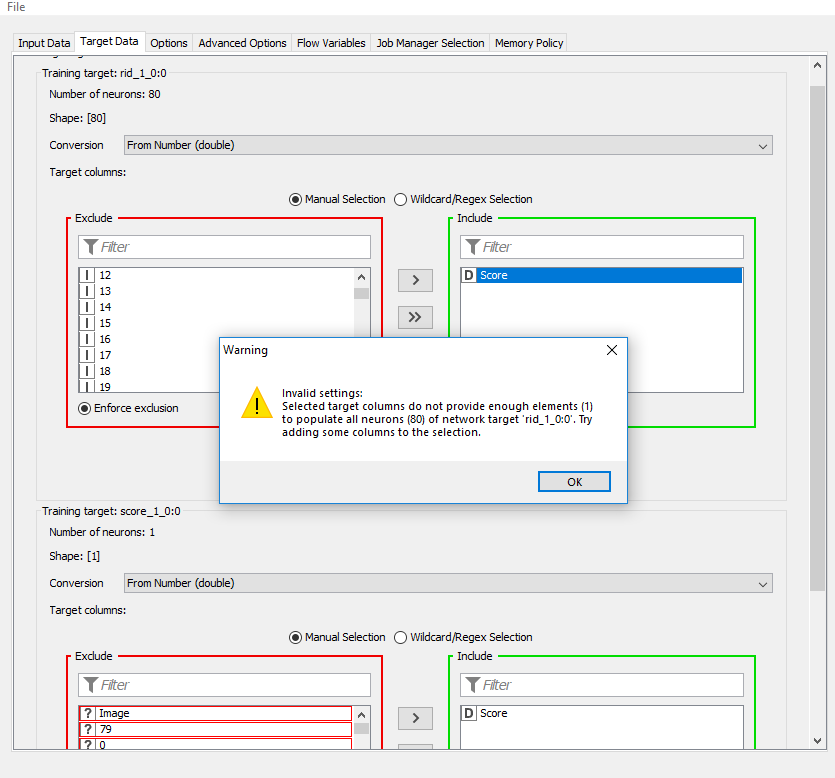
I gave another shot at this and I’m still coming up with errors; its now telling me I do not have enough “target elements”.
In the simple terms I want to build a model with (2) Inputs:
-
Image
-
RID Label
I want then want the output to be the Predicted Score (toxicity). I have labels for RID and Score, but Score is my target. I’m confused about if I should be using the score as an input or not.
Below are my Input and Output screenshots:
Is there another way to approach this? (to build a scoring model) I don’t want to give up on this, but when I hit these roadblocks, there is really no other option but to post here for help; I’m not a developer, just a researcher. I saw this blog, and thought maybe I could replace the classes with my categorical scores - https://www.knime.com/blog/using-the-new-knime-deep-learning-keras-integration-to-predict-cancer-type-from-histopathology
Thanks,
Anthony
Hello @asrichardson,
ok maybe I misunderstood what you are trying to achieve.
I thought you wanted to build a model that can predict both, the rid label and the toxicity score.
The workflow I provided above does that but their is still a small mistake in how you provide your targets.
For the first target (rid_1_0:0) you have to exclude the score and instead use the 80 integer columns that were created by the one-to-many node. They represent a one-hot encoding of your rid classification.
You should also switch the loss function from Binary cross entropy to Categorical cross entropy since I assume a sample can’t belong to multiple regions.
If you can provide me with a small subset of your images I can set up the workflow properly so you only have to run it.
Best,
nemad
Thanks for your quick reply @nemad and continued support & patience - I appreciate it.
Yes, the model requirements have changed - the RID will always be provided in the test set; Score is the constant unknown which I have to determine in the test set of 1K images.
A few more facts about the training set:
-
Consists of 2K~ images, for which the RID (region) and Score (toxicity) are labeled in a ground truth CSV.
-
There are 77 possible regions (RID) and 31 values in the range of toxicity scores (integers and decimal between 0 and 1) from which these 2K training images are labeled. (0 = no toxic cells present, vs. 1 = 100% toxic cellularity)
-
The images are based from about 32 different patients (for which a patient ID exists, but not needed at this time, since I need to be able to score on unknown patient images)
-
RID = the region from where the biopsy was taken from. That said, a toxic (.50 score) sample from let’s say RID #1 could look very different from an equivalent .50 toxicity sampled from RID # 77. (this can be due to the level of fat/muscle existing within or not, in a particular region).
I need to train a model, that can predict scores of any non-labeled images submitted to it, to give the highest probability of the Score. In the end, I need to be able to run inference on the model once complete, using random batches of images.
My advisor believes this can simply be achieved via a regression model; I thought it could be done via classification with the proper last layer and activation function. I am not certain on how to build this, because I have not observed any similar examples in the forums/posts and because I am new to the platform.
If you have any thoughts on how to build a regression model in KNIME, or if you observe a better way to tackle this, I am open to your ideas. I would like to create a project in the KNIME community, covering this use case, once I have success.
I have uploaded a directory to (100) images along with a CSV with labels; please find here: https://drive.google.com/file/d/1g1CfgeJzKfVH03vWUM-JOidkUWfmknSt/view?usp=sharing
Any support you can provide is greatly appreciated, thanks.
-Anthony
Hi @asrichardson,
I actually spend quite some time with your data but I had some troubles with my hardware which prevented me from creating a fully deep learning based workflow.
However, I also tried a different approach which also uses a deep learning model but only as feature extractor.
The rough idea is to feed the images to a deep learning model (e.g. InceptionResNet) which extracts a feature vector describing the images. These features are then combined with the region indicator and fed to a “shallow” learner e.g. the Gradient Boosted Trees Learner.
If you already updated to KNIME 3.7.0, you could also use the new XGBoost nodes which have the advantage of supporting logistic regression (the targets all lie in [0, 1]).
Here is a workflow that does what I described above: Keras_Shallow_Learning.knwf (89.4 KB)
Note that it uses the mentioned XGBoost nodes, so you will need KNIME 3.7.0 to use it.
If that’s not possible, you can simply replace the XGBoost nodes with Gradient Boosted Trees or another KNIME learner (and consequently predictor).
Best,
Adrian
Thank you for spending the time Adrian; I will this try this out today and follow-up when I have some results.
Hi Adrian, I finally got the model to run! This is some genius work here!!! I have a new reinvigorated mission to learn all I can about KNIME. I need to attend a KNIME training in the New England area if they are offered. If I could get this platform form in the hands of my undergraduate students, they would stick to it. @nemad
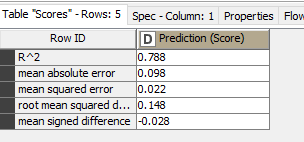
Is there anything you would recommend to increase performance? (batch size?, memory policy to full?)
I’d like to submit some unseen non-scored images (I only have the file name and RID info) to the trained model (for scoring) and check with my adviser on performance. Is there a simple way to run inference or deploy the model locally on my machine, so that I can just upload the images using the image reader? I do not want to disrupt the current architecture that was built. I’m betting money that I have to pre-process these new images - I was hoping to automate this step, but am willing to take the long path if you can share an example.
I’d like to include you as a contributor on a related paper I am preparing for 2019 (re: cementing the legitimacy of visual programming for machine learning). Will come back to this once I finish my semester.
Thanks,
Anthony
Hi Anthony,
I am glad to hear that the workflow fulfills its purpose 
If we can help you getting your students started, please let us know, maybe we can find a solution together.
Regarding Deployment:
To be honest, I didn’t think about deployment when I created the workflow, so I reworked it a bit to include both training and deployment: Keras_Shallow_Learning.knar (130.2 KB)
I had to exclude the images due to a file size limit here in the forum but if you run training on your machine you should use all your data not just the subset you sent me anyway.
This is because a) more data is usually better and b) not all rids are included in the subset so the model will fail on unseen rids.
Regarding your performance questions:
The main bottleneck is the feature extraction using InceptionResNetV2 mainly because this network is quite large. Theoretically, you could plug-in any pretrained network that extracts features from RGB images.
A good source for such networks is the keras.applications package shipped with keras.
A smaller network will run faster but the features it extracts might be not as good as those extracted from InceptionResNetV2. I have to admit that I was suprised that the features work so well (or rather at all) for your use case because the images the network was trained on (Imagenet) are from a completely different domain (natural images).
If you could get your hands on a network that was trained on similar images like the ones you have, I’d expect that the results would be considerably better than what you already get now.
Best,
Adrian
Thank you, Adrian, yes, I can certainly talk more about classroom integration after 12/20 (official semester end).
Thank you for the revised model; I will try later today and let you know how it works. This entire process makes me realize the benefit of running a KNIME server with an Agile team - the collaboration potential is endless.
Yes, I did, in fact, use all the ~2K training images to train the model and extract the features; the feature extraction took about 90mins - my CPU was at 90-100%, but I have 20GB of RAM. I have a 2GB GPU; not sure if that could be utilized with KNIME and the DL nodes. I’m guessing it depends on how I configure Anaconda and Keras Extension for the Python Integration.
Thanks for the suggestions about performance; are those other networks you suggest (via keras.applications package) accessible via the KNIME Keras extension? @nemad
Thanks,
Anthony
Yes that’s the idea behind the server.
You could try to increase the batch size but tensorflow is not exactly economical with its memory usage so you might find that your 20GB are filled up quickly (also make sure that you increase the memory KNIME can access in the knime.ini file).
Usually GPUs speed up deep learning related tasks (both training and deployment) by quite a bit but you will have to check whether your GPU is compatible with CUDA 9. If it is then Anaconda will typically perform the setup for you (we are currently updating our documentation and it will then hopefully be much easier to setup python for deep learning).
They are in the same way as InceptionResNetV2, so you have to write a bit of python script similar to the one included in the DL Python Network Creator node in the Training workflow.
Depending on the network you will have to adjust the constructor call but everything you need to know is documented here: https://keras.io/applications/
Many of these networks use the same preprocessing but if the model with the new network doesn’t work at all, it might be because the new network requires a different preprocessing.
Best,
Adrian
This topic was automatically closed 90 days after the last reply. New replies are no longer allowed.