Hi,
I have 400 MB size text file (About 1M rows of data and 85 columns) that I am reading from an S3 location using the Python source node.
I use pd.read_csv function to read the file with the below arguements. I understand converters increase the time taken to parse the column type. If I don’t specify that, the node fails with the message ‘column ‘X’ doesn’t support type long’ etc.
df = pd.read_csv(BytesIO(res), delimiter = “|”, error_bad_lines=False, converters = {int: str})
With the above, it takes an hour to read the file. The columns are parsed correct.This serves as an input data to a subsequent workflow in KNIME. I want to be able to run this is in the server and that’s why I am using S3 and automating the workflow.
It I read the file using Jupyter notebook, the same file takes less than 5 minutes to read.
How can I optimise the file read in KNIME?
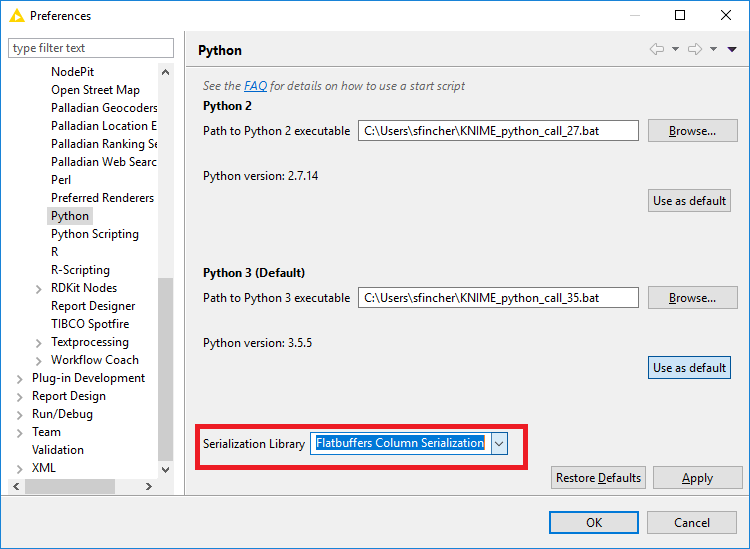
One thing you can try would be to change the transfer library in KNIME’s Python preferences from Flatbuffers to Apache Arrow. I know this has improved performance for at least one other user - not certain if it will work in your case, but it’s an easy test and worth a shot!
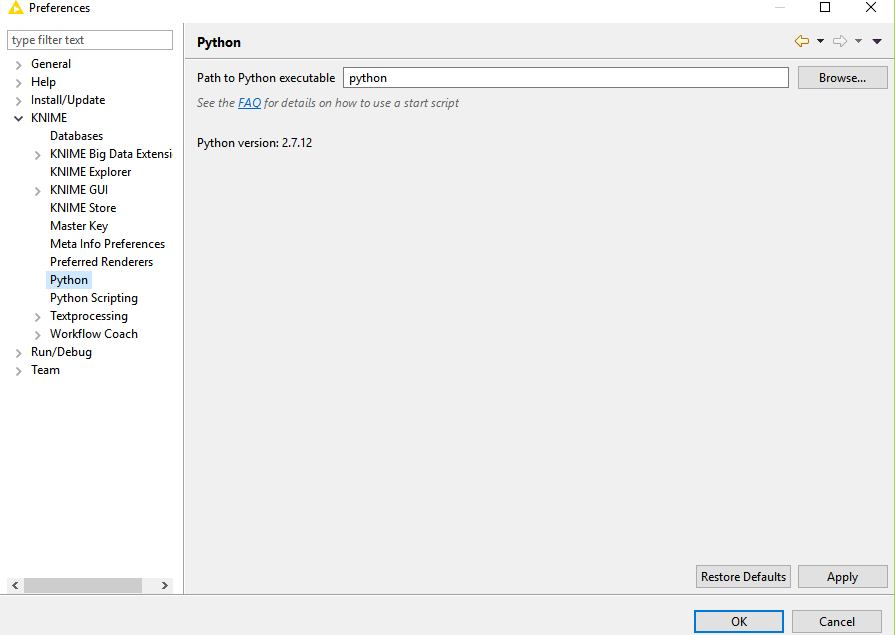
I am using KNIME version 3.3.2. I don’t see the serialisation option in my preferences. Screenshot attached.
We rewrote the Python extensions in KNIME 3.4, so you might consider an upgrade, if that’s possible. I believe the serialization library options are only available in newer versions of KNIME.
If you do decide to upgrade, we have a helpful post about how to setup Python with KNIME here: https://www.knime.com/blog/setting-up-the-knime-python-extension-revisited-for-python-30-and-20
We are using KNIME server 3.4 for now in my company. I was advised to use the desktop version 3.3.2 so that it is compatible. We are planning to upgrade the KNIME server soon and then I will update the desktop version. I shall try this out then again.