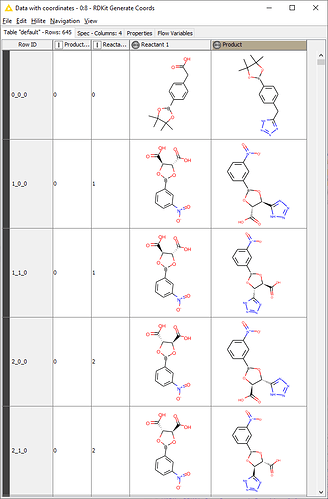
SMARTS strings are very tricky. I use ChemDraw and this SMARTS tool to generate my SMARTS strings (though it can’t do reactions). It also takes a lot of trial and error and referring back to the SMARTS documentation.
In your specific case, the string [CX3:1](=O)[OX2H1]>>[CX3:1]1=NN=NN1 gets results that look decent.
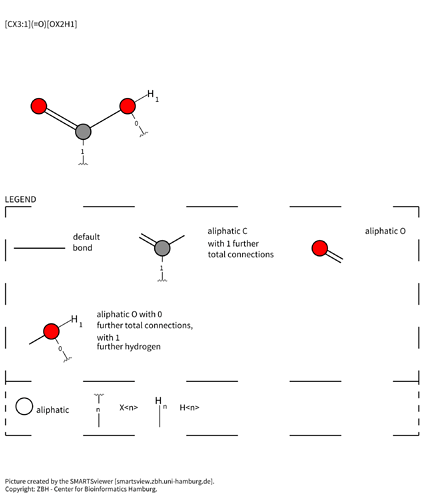
[CX3:1](=O)[OX2H1] is the carboxylic acid with the carbon mapped as atom 1:
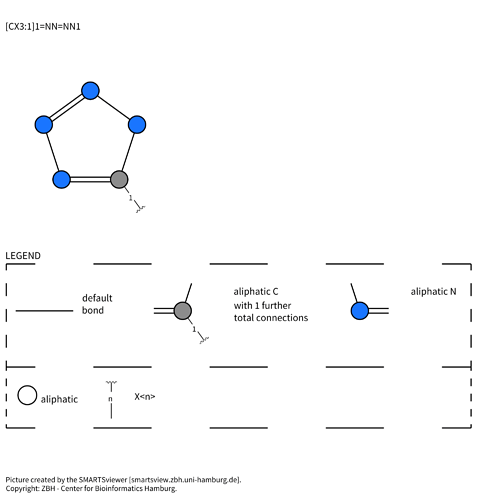
[CX3:1]1=NN=NN1 is the tetrazole with the carbon mapped as atom 1:
The mappings tell the reaction node which positions to act on.
Finding a simple automated way to do this would be great, but I haven’t come across one. I’d be curious to know if anyone else has.