Hello everyone,
I am using the ImageJ extension in KNIME to do some chromatin image analyses. The goal is to measure both the nucleus volume and chromatin volume of a cell. I have managed to use simple thresholding to generate nucleus and chromatin binary images. Eventually I will be doing this on a field image of multiple cells so I have also used Trackmate tracker to track the nuclei and chromatin.
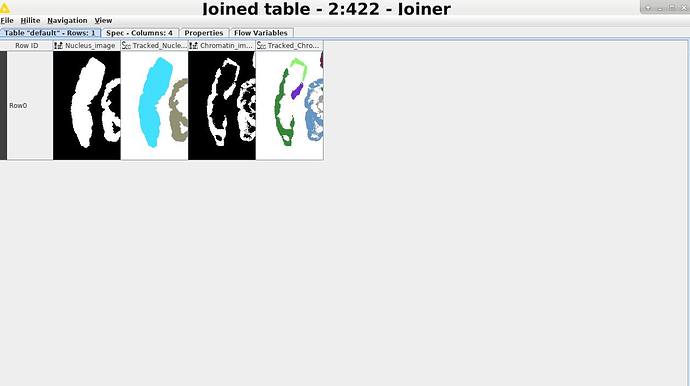
Here is a table where I have the nucleus image and tracking (label) and the chromatin image and tracking. For now, please focus on the nucleus on the left.
My issue is that the chromatin stain within a nucleus can be discontinuous (a feature of the biological phenomenon I’m studying), and therefore identified by the tracker as separate objects. Since my goal is to have the nucleus and chromatin volume of a cell, how do I make KNIME recognise that, say, chromatin objects 1, 2, 3, all belong to the same nucleus (e.g. nucleus 1)?
I have tried using Image Segment Features, using nucleus track for ‘Labelling’ and the thresholded binary chromatin image for ‘Image’, selecting numpix in Features, and checking the boxes for ‘Append labels for overlapping segments’ & ‘Overlapping segments do not need to completely overlap’. However, the result in ‘Calculated Features table’ is still the nucleus volume, not the chromatin volume.
Based on a this thread (Feature Calculator - #5 by gab1one) it seems that I’m using the right node for what I want to do. Would anyone have any suggestions?
Thank you!