Dear all,
I have a workflow and need to account for a column which can either be a String column or an RDKit Molecule column.
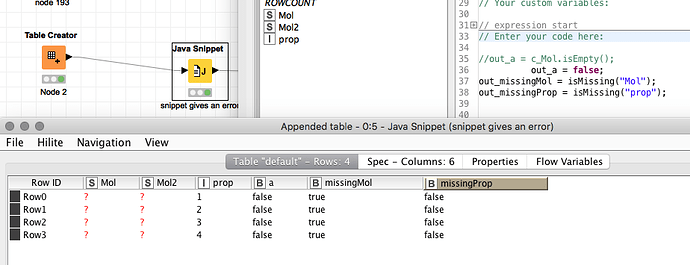
What I know is that if the column is empty, it is a string column, otherwise it is an RDKit column.
I came up with two ideas:
a) Check if column is empty:
Apporach: use the function which is proposed by the Java Snippet node:
c_column.isEmpty()
Unfortunately this does work only if the column is a String, but not for the RDKit Mol column.
I could try it and catch the exception but that seems a bit weard.
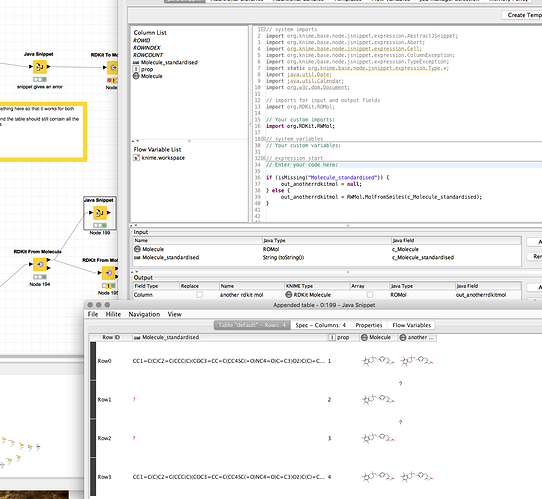
b) Check if the column is of type String:
Approach: use the isType(name, type)* function as described here:
isType(“MyColumn”, tString)
Yet, that always returns true, so I guess I somehow have to enter the Column name differently, but I have no clue how.
If I input the column itself I get the error
“The method isType(java.lang.String, T) in the type org.knime.base.node.jsnippet.expression.AbstractJSnippet is not applicable for the arguments (org.RDKit.ROMol, java.lang.String)”
c) The nodes I have also tried so far:
The table Validator fails because he cannot cast a String cell to an rdkit cell.
The extract table spec is an option but somehow its hacky and I hope there is a more elegant solution.
The cherry on the cake would be if I could cast the String column to RDKit and be done…but I would be also glad if I could somehow get solution a or b working somehow (with a node, some code snippet or whatever works)
It would be glad to get some help here.
Thanks in advance,
Jenny
PS: I have not posted it to the RDKit Forum since I think that this task could also hold true for other column types which need to be casted and/or checked.