Hi all,
my question is maybe a bit naive. I was wondering if there is some way to download the latest release of ChEMBL database (with data about compound IDs, smiles, bioactivity data) by using knime nodes, or if you could direct me to some exemplary workflows. I saw that most of the workflows allow to extract a limited number of compounds or compounds active on a given target. Maybe some database nodes?
Thank you in advance
Hi @C_C_85
It is possible to download the whole ChEMBL database in different database formats from the following gitbook:
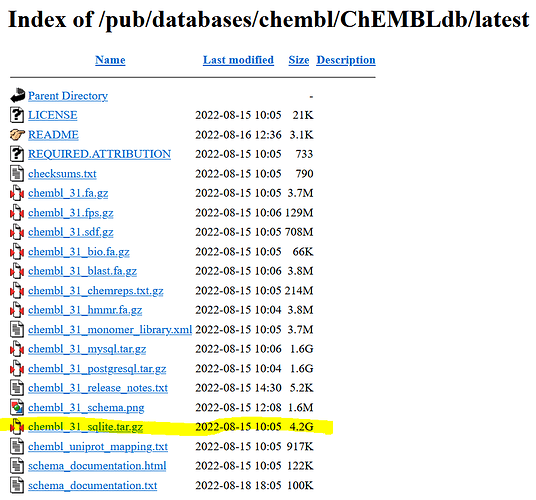
To make the installation in KNIME the simplest, I would recommend to download the SQLite which is an embedded database and therefore does not need the installation of a database server:
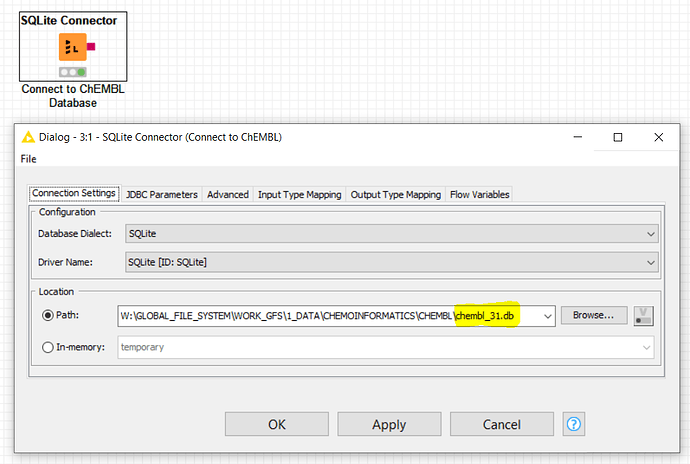
Once it is downloaded, you would just need to unzip it and untar it, so that it can be directly referenced and accessed by the KNIME -SQLite Connector- node:
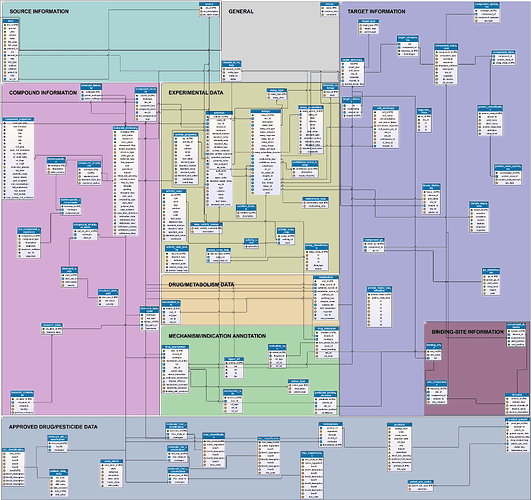
In the same repository as above, you will also find the schema documentation text file and the html web page. They list and explain all the tables and relationships of the ChEMBL database:
![]() schema_documentation.html 2022-08-15 10:05 122K
schema_documentation.html 2022-08-15 10:05 122K
![]() schema_documentation.txt 2022-08-18 18:05 100K
schema_documentation.txt 2022-08-18 18:05 100K
Moreover, all the molecules and substances referenced in ChEMBL can be downloaded separately from the same repository above. They are stored in file
![]() chembl_31.sdf.gz 2022-08-15 10:05 708M
chembl_31.sdf.gz 2022-08-15 10:05 708M
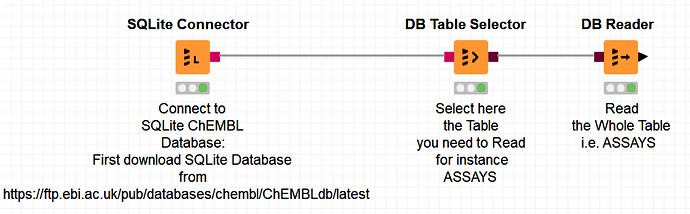
The following workflow is a very simple example of how to access the SQLite ChEMBL database:
It uses the KNIME -SQLite Connector- node to connect to the SQLite ChEMBL database in a first instance, as follows:
and then reads one of the tables, as follows:
The downloading and reading of new versions of the SQLite ChEMBL database could be automated using other KNIME nodes but its manual installation in this way is so simple that maybe it suffices for the time being ?
Hope all these hints help to answer your question.
Best
Ael
Thank you so much for your suggestions and providing the workflow, it is a great place to start!
Is it possible to select more than one table using the node DB Table selector? I have no experience building sql queries; after these steps, i need to map compounds and activities relevant informations.
Hi @C_C_85
Glad it helped you to start with ChEMBL databases. Definitely it is possible to open/read several tables at the same time.
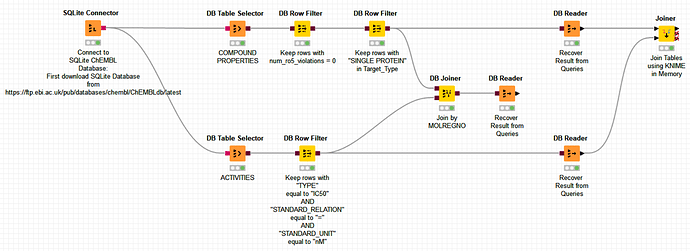
The beautifulness of KNIME is that one does not need to know SQL to do queries. To start querying the database, you can use equivalent SQL nodes to those existing in KNIME. For instance, -DB row Filter-, -DB Column Filter- and -DB Joiner- nodes:
I’m adding here a dummy example of workflow which achieves different SQL queries (without having to write them by hand :)):
The final query is joining two different ChEMBL tables, one for Compounds and a second one for Targets to gather activities from compounds on targets.
Hope it helps.
Best
Ael
Ps: By the way, there is an image of the ChEMBL schema available from here which is quite helpful to understand the relationships between the different database tables:
Hi @C_C_85
Did the last workflow example help you to query the ChEMBL database and read several tables? Your feedback will be more than welcome.
Best
Ael
Hi, sorry for not getting back to you sooner.
The workflow example is great, thank you so much! I think that many other users will certainly benefit from it!
Best
C.
Hi @C_C_85
No problem at all and glad you liked it. As you said, I hope too it will help other people from the forum too.
Thank you for validating the solution !
Best wishes,
Ael
This topic was automatically closed 7 days after the last reply. New replies are no longer allowed.