let’s see if I understand correctly.
Each row of the dataset represents a case, which occurred in a certain year / month in a certain municipality. In the same row there is the population of the municipality, identical in each row that refers to that municipality.
We want to calculate, for each municipality, year and month
- the incidence = (number of cases in the month / municipality population) * 10,000
- hospitalization rate
- lethality rate
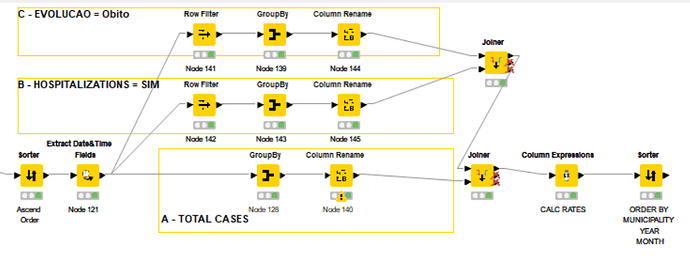
A - First we have to count the total cases in each municipality in each month of each year. To do this we use the Groupby node and we group by year, month and municipality.
To count the number of rows we use a trick: we count the number of values present in any column, for example Reg_COVID. The population of each municipality is always the same, so we take the first value in the group (we could take the max or min value as well)
B - In a parallel branch we filter the dataset in order to obtain only the “SIM” and then group in the same way, this time omitting the population
C - In a parallel branch we filter the dataset in order to obtain only the “Obitos” and then we group as in B
D - We combine the three tables. We now have all the data we need to calculate the rates
E - We calculate the three rates and rearrange the resulting dataset
This is the final part of the workflow
This is the result (part of)
KNIME_covid_brazil.knwf (124.0 KB)