Hello,
Thank you.
Can you post your workflow and dataset so that we can recreate the error?
Here is the workflow and a sample of the data
Workflow
Sample of Data
<cas.rn>
<cas.index.name>
<molecular.formula>
<molecular.weight>
<boiling.point.predicted>
<density.predicted>
<pka.predicted>
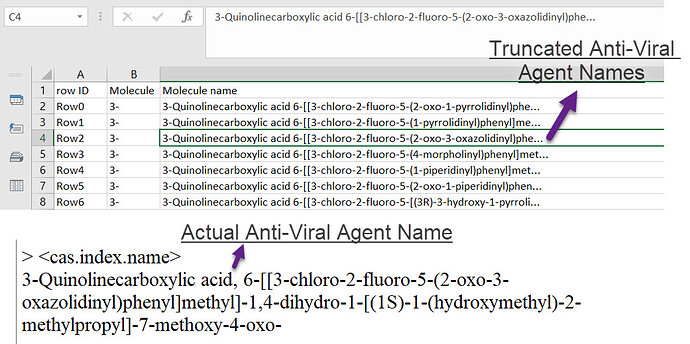
Just reading the excerpt you posted from the sd file, the text in the molecule name field is already truncated from the start (see the first line).
Assuming you didn’t create the initial sd file, there’s nothing you can do about that, and I’d argue that it’s the least important part of the file anyway.
What does the cas.index.name column look like?
In the future, posting a workflow that we can open ourselves, instead of just a screenshot, would be more helpful.
2 Likes
The first is truncated but the ones that follow that aren’t truncated in the sdf file are further shortened in the file excel file.
The workflow folder is around 60MB. how do i post the workflow over here?
You can upload workflows to your KNIME Hub space: https://hub.knime.com/site/about
1 Like
system
September 28, 2020, 7:17pm
8
This topic was automatically closed 7 days after the last reply. New replies are no longer allowed.