Hi Everyone,
I have an SD file from a vendor partner and they have errors in the formatting which do not allow the correct reading of the SD file. In this specific example there are two carrage return and line feeds between the molecule end (M END) and the first token which is the catalog number.
I’m looking for any help on how others in the community may have corrected these errors using KNIME as the number of errors are large and the file is also large so manually is not sustainable. I will need to be processing this file on a routine basis and unfortunately the vendor does not have the knowledge to correct their file.
My goal is to create a workflow which can solve these types of issues and post it on the HUB for others. Currently I don’t see any content but I don’t believe I’m the only one dealing with these types of issues 
Thanks in advance,
Jason Ochoada
could you share part of that file?
you could text edit it and only keep the first 10 or so compounds? Assuming you know the structure of an sdf file.
I have to dig through some archived workflows, but I think I stumbled upon a similar problem many moons ago - something about using the line-reader node within a loop and correcting (the line ending) with regex replace or such.
It’s possible that if the file isn’t that big that you can solve it though faster within e.g. notepad++ by reading the whole file, visualizing hidden tokens and using find/replace. But yes, that’s not Knime 
2 Likes
Certainly! Thanks for being willing to take a look. I have attached the full file here. HY-L022P.zip (2.0 MB)
Agree with @docminus2 Using notepad++ or similar text editor for initial cleaning is probably easiest not automated. What worked for me with the attached file:
There are many occurrences of 3 line breaks in succession. This is invalid. They need to be replaces with 2 line breaks instead. I did this in notepad++:
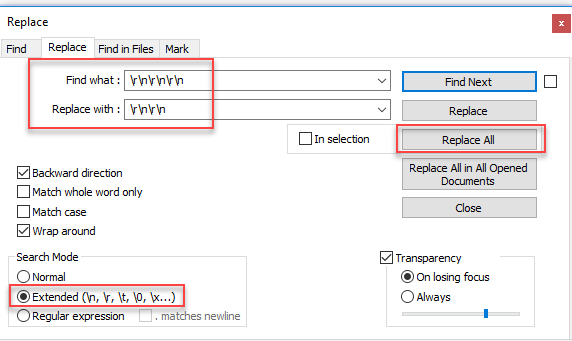
SDF Reader still has issues with about 30 entries but they seem to contain html and weird chars in the properties.
I also tried the Vernalis SDF Reader (Load SD-Files). This reads everything and with the SDF Extractor you can then get the properties. However getting the properties also fails for the same 35 molecules but you at least get the structure (maybe that is enough?).
All in all the real solution is to tell the supplier to send valid usable sd-files…
EDIT: It seems one needs to replace even more of the too many line breaks. There are up to 5 successive breaks in some of the properties. In other cases there are also just multiple new lines followed by line breaks. It’s a mess. Only supplier can really fix this IMHO.
2 Likes
@docminus2 @beginner Thank you both so much for taking a look at this. Looks like the remaining issues are missing headers.
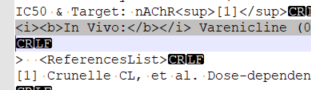
Yes I agree that the vendor should fix this but in reality I don’t believe they have the capability.
Now maybe with some string manipulation I can fix this file in KNIME and release it on the hub. Let me know if you have any additional ideas.
Thanks so much again!
Jason



