So I have a workflow that reads in a list of files with “List Files/Folders” node, then in an R snippet reads each of the files and binds all of the rows and appends unique columns.
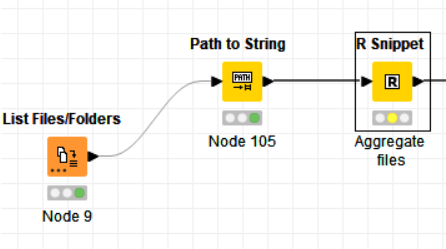
The code in the R snippet is:
library(tidyverse)
library(dplyr)
library(tidyr)
library(janitor)
fil_locat<- “C:\Docs\Data Science\Refresh_2024-03-20”
files_load ← list.files(path = fil_locat, pattern = “*.csv”, full.names = TRUE)
df ← files_load %>% map_df(~read_csv(.,col_names=TRUE,id=‘path’, show_col_types = FALSE)) %>% bind_rows() %>% clean_names() #%>% as.data.frame()
write.csv(df,file=“C:\Docs\Data Science\Data\results_r.csv”, row.names=FALSE)
knime.out ← df %>% as.data.frame()
While in the node, when I inspect either df or knime.out, the column cohort_day shows a date and shows to be of type date.
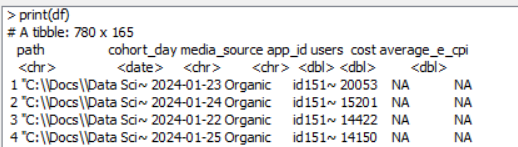
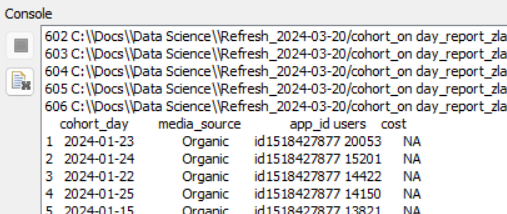
But examining the data Output of the node it shows “cohort_day” is now a double? With max and min values of 19799 and 19723 respectively which are typical excel date integers.
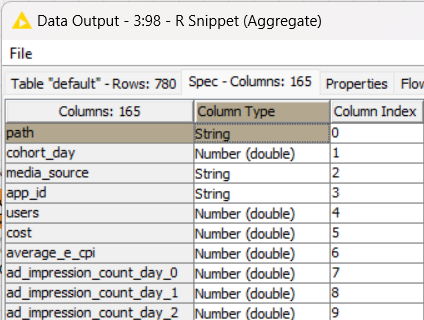
I guess I can convert them and write them back, but why the difference inside the node compared to what is being outputted by the node?