kNIME DEEP LEARNING Mutagenicity.xml (756.7 KB)
Hi
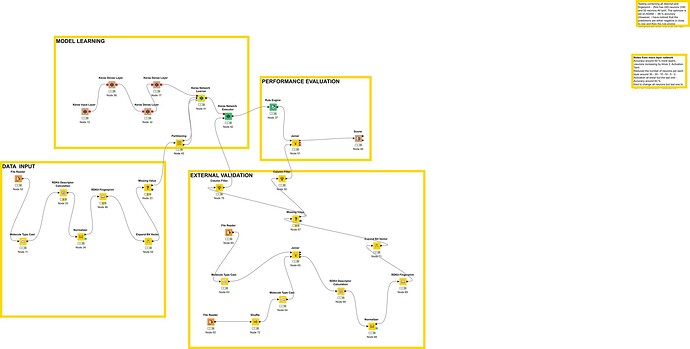
I have created a deep learning workflow to predict mutagenicity (BINARY value) in drugs impurities.
The workflow has been created combining existing nodes from various extension.
The models takes molecules smiles string and construct predictors (molecular fingerprint, and chemical properties combined )which are then mathematically associated with mutagenicity by the KERAS deep learning model. The model has been tested with an external database and its accuracy compared with DEREK ( 86% vs 90% of DEREK)
In pharmaceutical drugs there are certain impurities that are present in very low concentration and almost impossible to isolate. This difficulty associated with in vitro testing allows ,nowadays, computational methods to predict the mutagenicity response which are valid for the legislative purposes (FDA approval).
Thanks
kNIME DEEP LEARNING Mutagenicity.json (638.2 KB)
ttached the JPG, JSON, and SVG of the workflow