Hi All, I'm having the same issue too.
I have KNIME 3.5.1, have already set the R buffer size to unlimited.
At first, if I evaluate the script (which works perfectly in RStudio), all goes well, up until the point, where I try using the nmf from the NMF package. Then I get the error message from above, which is:
ERROR: R evaluation failed.: "knime.tmp.ret<-NULL;printError<-function(e) message(paste('Error:',conditionMessage(e)));for(exp in tryCatch(parse(text=knime.tmp.script),error=printError)){tryCatch(knime.tmp.ret<-withVisible(eval(exp)),error=printError)
if(!is.null(knime.tmp.ret)) {if(knime.tmp.ret$visible) print(knime.tmp.ret$value)}};rm(knime.tmp.script,exp,printError);knime.tmp.ret$value"ERROR: R evaluation failed.: "knime.tmp.ret<-NULL;printError<-function(e) message(paste('Error:',conditionMessage(e)));for(exp in tryCatch(parse(text=knime.tmp.script),error=printError)){tryCatch(knime.tmp.ret<-withVisible(eval(exp)),error=printError)
if(!is.null(knime.tmp.ret)) {if(knime.tmp.ret$visible) print(knime.tmp.ret$value)}};rm(knime.tmp.script,exp,printError);knime.tmp.ret$value"ERROR: eval failedERROR: Could not capture output of command.ERROR: R evaluation failed.: "sink();sink(type='message')
close(knime.stdout.con);close(knime.stderr.con)
knime.output.ret<-c(paste(knime.stdout,collapse='\n'), paste(knime.stderr,collapse='\n'))
knime.output.ret"ERROR: R evaluation failed.: "sink();sink(type='message')
close(knime.stdout.con);close(knime.stderr.con)
knime.output.ret<-c(paste(knime.stdout,collapse='\n'), paste(knime.stderr,collapse='\n'))
knime.output.ret"ERROR: eval failed, request status: error code: 127ERROR: Could not execute internal command.ERROR: R evaluation failed.: "dev.off()"ERROR: R evaluation failed.: "dev.off()"ERROR: eval failed, request status: error code: 127
After which all objects are forgotton, knime.in not found, functions not found, everything not found, even though the objects are still listed on the right.
Any ideas? (I'm happy to send the wf, if you'd like to take a look.)
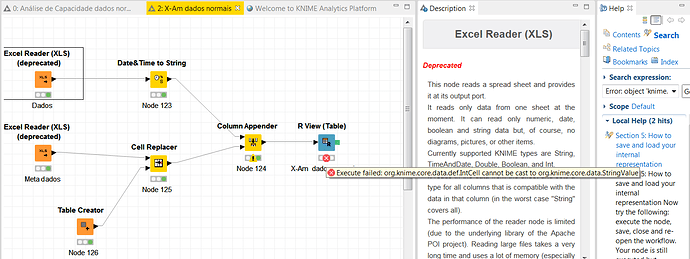
 If anything, the R View node is not generating a useful error message to help diagnose the problem. I think this behavior is probably a bug, so I’ll run it by one of our developers for clarification.
If anything, the R View node is not generating a useful error message to help diagnose the problem. I think this behavior is probably a bug, so I’ll run it by one of our developers for clarification.