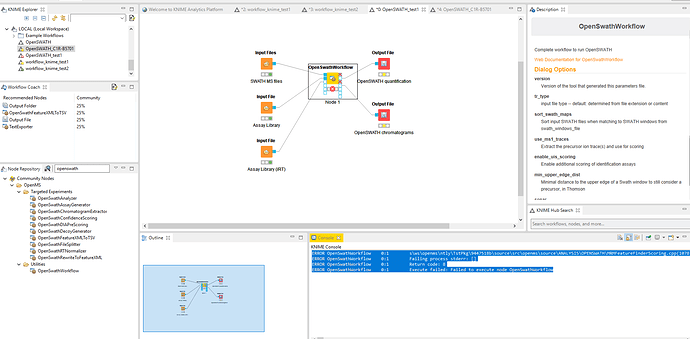
Hello All,
I am using KNIME to run OpenSWATH (OpenMS v. 2.5.0) to analyze my DIA MS data. I have utilized the shared workflow for OpenSWATH (as a screenshot attached) but encounter some errors when I organize everything as recommended in the manual tutorial. The errors are here:
ERROR OpenSwathWorkflow 0:1 s\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 39 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 40 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 41 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 42 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 43 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 44 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 45 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 46 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 47 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 48 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 49 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 50 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 51 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 52 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 53 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 54 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 55 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 56 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 57 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 58 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 59 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 60 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 61 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 62 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 63 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 64 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 65 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 66 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 67 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 68 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 69 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 70 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 71 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 72 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 73 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 74 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 75 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 76 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 77 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 78 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 79 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 80 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 81 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 82 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 83 from group 0 does not have a corresponding chromatogram, C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMFeatureFinderScoring.cpp(1078): Error: Transition 84 from group 0 does not have a corresponding chromatogram, Will analyse 0 peptides with a total of 84 transitions , C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\OpenSwathWorkflow.cpp(191): Extracted best features: 0, Error: Unexpected internal error (Need at least 3 data points to remove outliers for the regression.), Error occurred in line 141 of file C:\jenkins\ws\openms\ntly\TstPkg\9447518b\source\src\openms\source\ANALYSIS\OPENSWATH\MRMRTNormalizer.cpp (in function: class std::vector<struct std::pair<double,double>,class std::allocator<struct std::pair<double,double> > > __cdecl OpenMS::MRMRTNormalizer::removeOutliersIterative(const class std::vector<struct std::pair<double,double>,class std::allocator<struct std::pair<double,double> > > &,double,double,bool,const class std::basic_string<char,struct std::char_traits,class std::allocator > &)) !]
ERROR OpenSwathWorkflow 0:1 Failing process stderr:
ERROR OpenSwathWorkflow 0:1 Return code: 8
ERROR OpenSwathWorkflow 0:1 Execute failed: Failed to execute node OpenSwathWorkflow
I would be appreciated if you guide me on how can I fix it. Alternatively, you can contact me via my email address: mohammad.shahbazy@monash.edu
Thank you in advance,
Mohammad