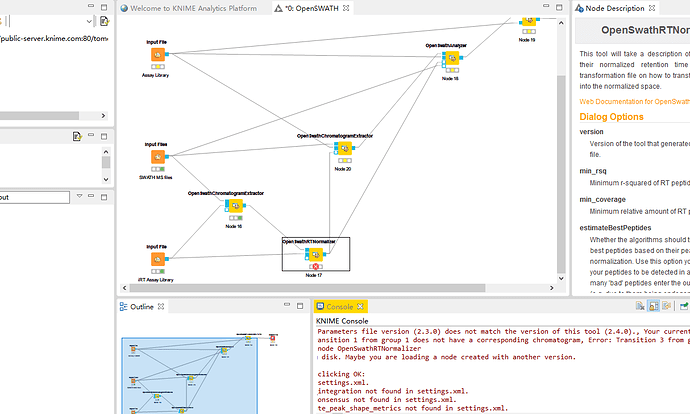
When I run this module“OpenSwathRTNormalizer”,There was an error,Warning: Parameters file version (2.3.0) does not match the version of this tool (2.4.0)., Your current parameters are still valid, but there might be new valid values or even new parameters. Upgrading the INI might be useful.I know my openMS is 2.4.0,but what is the Parameters file version (2.3.0),how do I upgrade it,what is the INI,where are they.
Hi,
the warning about the parameters can be ignored. It just tells you that the workflow was created with an earlier version of the OpenMS plugin. You can solve it by dragging the node freshly in there (edit: but careful, you will lose the settings).
I would be worried about what the error tells you next.
Error: Transition 3 ...
Best
Julianus
Hi‘
Thank you for your reply,here are the next errors,and I don’t know how to deal with it.
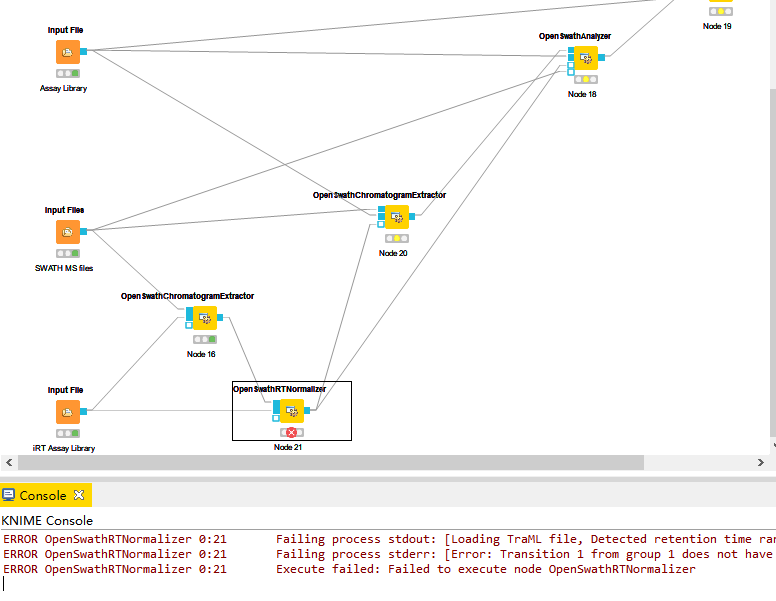
ERROR OpenSwathRTNormalizer 0:21 Failing process stdout: [Loading TraML file, Detected retention time range from -28.3083 to 86.7152, RT Normalization working on C:\Users\mazhen\AppData\Local\Temp\knime_OpenSWATH62023\fs-OpenS_0-16-62024\000\000\OpenSwathChromatogramExtractor_0\split_napedro_L120420_010_SW-400AQUA__human_2ul_dilution_1_0.nf.pp.mzML, Progress of ‘loading chromatogram list’:, , – done [took 0.00 s (CPU), 0.00 s (Wall)] – , Will analyse 2 peptides with a total of 56 transitions , Error: Unexpected internal error (Need at least 3 data points to remove outliers for the regression.)]
ERROR OpenSwathRTNormalizer 0:21 Failing process stderr: [Error: Transition 1 from group 1 does not have a corresponding chromatogram, Error: Transition 3 from group 1 does not have a corresponding chromatogram, Error: Transition 4 from group 1 does not have a corresponding chromatogram, Error: Transition 6 from group 1 does not have a corresponding chromatogram, Error: Transition 11 from group 2 does not have a corresponding chromatogram, Error: Transition 12 from group 2 does not have a corresponding chromatogram, Error: Transition 13 from group 2 does not have a corresponding chromatogram, Error: Transition 14 from group 2 does not have a corresponding chromatogram, Error: Transition 15 from group 2 does not have a corresponding chromatogram, Error: Transition 17 from group 2 does not have a corresponding chromatogram, Error: Transition 19 from group 2 does not have a corresponding chromatogram, Error: Transition 31 from group 4 does not have a corresponding chromatogram, Error: Transition 34 from group 4 does not have a corresponding chromatogram, Error: Transition 35 from group 4 does not have a corresponding chromatogram, Error: Transition 41 from group 5 does not have a corresponding chromatogram, Error: Transition 42 from group 5 does not have a corresponding chromatogram, Error: Transition 43 from group 5 does not have a corresponding chromatogram, Error: Transition 44 from group 5 does not have a corresponding chromatogram, Error: Transition 48 from group 5 does not have a corresponding chromatogram, Error: Transition 51 from group 6 does not have a corresponding chromatogram, Error: Transition 53 from group 6 does not have a corresponding chromatogram, Error: Transition 55 from group 6 does not have a corresponding chromatogram, Error: Transition 56 from group 6 does not have a corresponding chromatogram, Error: Transition 57 from group 6 does not have a corresponding chromatogram, Error: Transition 61 from group 7 does not have a corresponding chromatogram, Error: Transition 62 from group 7 does not have a corresponding chromatogram, Error: Transition 63 from group 7 does not have a corresponding chromatogram, Error: Transition 64 from group 7 does not have a corresponding chromatogram, Error: Transition 65 from group 7 does not have a corresponding chromatogram, Error: Transition 66 from group 7 does not have a corresponding chromatogram, Error: Transition 67 from group 7 does not have a corresponding chromatogram, Error: Transition 68 from group 7 does not have a corresponding chromatogram, Error: Transition 70 from group 7 does not have a corresponding chromatogram, Error: Transition 81 from group 9 does not have a corresponding chromatogram, Error: Transition 82 from group 9 does not have a corresponding chromatogram, Error: Transition 83 from group 9 does not have a corresponding chromatogram, Error: Transition 85 from group 9 does not have a corresponding chromatogram, Error: Transition 86 from group 9 does not have a corresponding chromatogram, Error: Transition 102 from group 11 does not have a corresponding chromatogram, Error: Transition 103 from group 11 does not have a corresponding chromatogram, Error: Transition 104 from group 11 does not have a corresponding chromatogram, Error: Transition 108 from group 11 does not have a corresponding chromatogram, Error: Transition 109 from group 11 does not have a corresponding chromatogram, Error: Transition 110 from group 11 does not have a corresponding chromatogram]
ERROR OpenSwathRTNormalizer 0:21 Execute failed: Failed to execute node OpenSwathRTNormalizer
Hi,
I am not an expert in the OpenSWATH part of our toolset but I know that the usage of OpenSWATH was eased by the new tool/node called OpenSWATHWorkflow, which combines most of the nodes which you are currently trying to use into one.
You can have a look at a recent tutorial that covers this.
Specifically section 6.5 but I recommend to read the Introduction and the full OpenSWATH part.
Also there are great instructions for the tools in the OpenSWATH environment here and here.
If you still have specific questions after this, I would advise you to send an email to the mailing list
https://sourceforge.net/projects/open-ms/lists/open-ms-general
There are more OpenSWATH experts listening.
Best,
Julianus
Thanks,
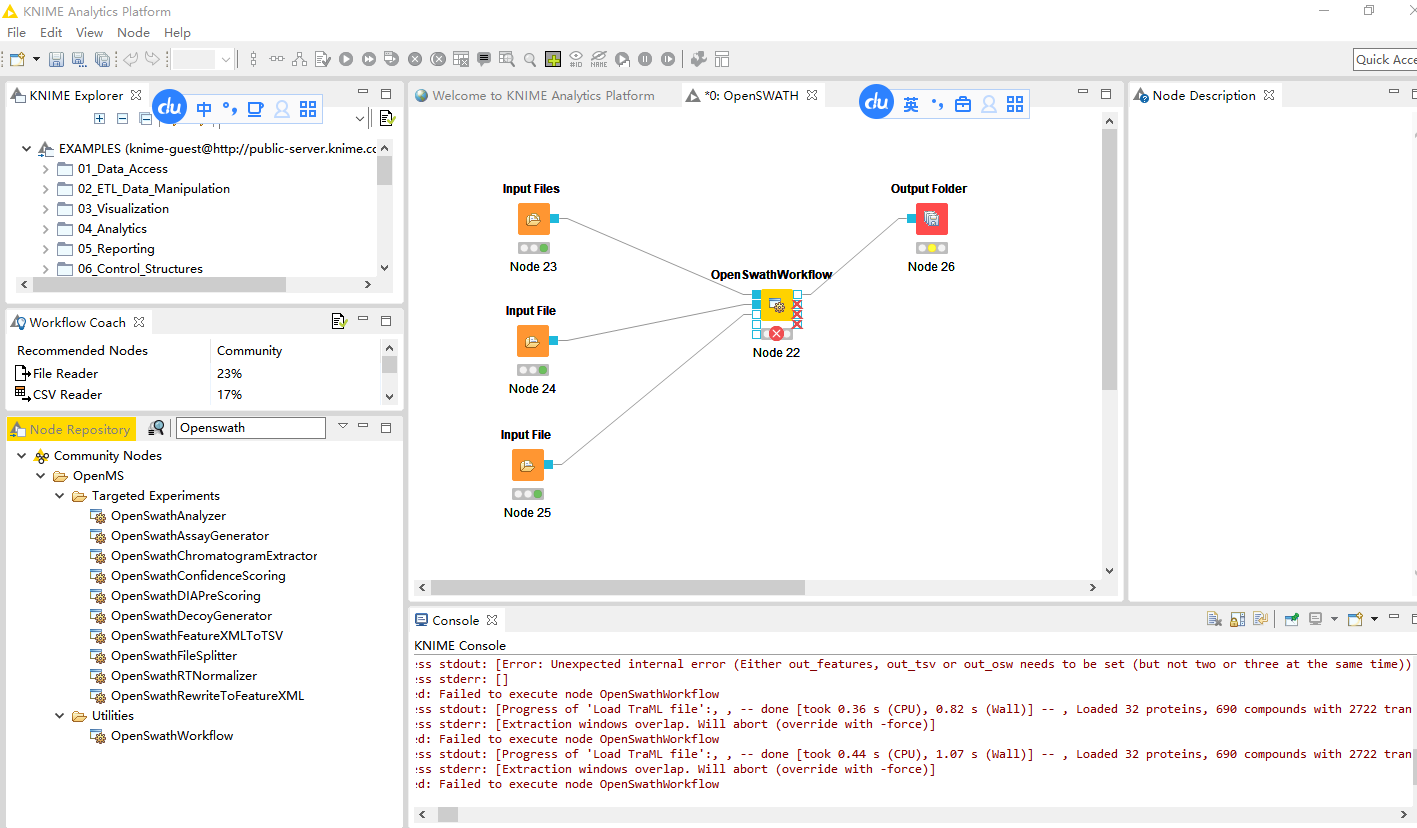
I have read the tutorial you send me,and I use the example datas and the node named Openswathworkflow provided by KNIME, here are still some errors,I just wanna know why and how to deal with it.
Extraction will overlap between 425 and 424.5, This will lead to multiple extraction of the transitions in the overlapping regionwhich will lead to duplicated output. It is very unlikely that you want this., Please fix this by providing an appropriate extraction file with -swath_windows_fil
I have logged into this website"https://sourceforge.net/projects/open-ms/lists/open-ms-general",but am not familiar with this, How can I get the effective contact information.Could you give me some contact informations for the developers
Regards,
Mazhen
The second questios is that swathdata always include MS2 data,why the example datas only include MS1 datas.
Hi,
the link for the mailing list is to sign up for news and help and to read old posts.
To post to the mailing list please use: open-ms-general at lists . sourceforge . net
The workflow is not connected the same as in the tutorial (it uses the first instead of the second and third output) so it might not work correctly.
But please post to the mailing list what kind of data you have and what you actually want to do and I am sure you will receive a good answer.
Cheers
Julianus
This topic was automatically closed 90 days after the last reply. New replies are no longer allowed.