I’m using Python to develop a KNIME extension, and I’d like to use the KNIME Base Chemistry types. I’ve made two of the nodes so far - but I get this strange error when I try to run the second one:
ERROR Molecule Properties 3:3591:3592 Execute failed: An error occurred while calling o17.getDataSource.
: java.lang.ClassCastException: class org.knime.chem.types.AbstractStringBasedValueFactory$AbstractCellReadValue cannot be cast to class org.knime.chem.types.MolValue (org.knime.chem.types.AbstractStringBasedValueFactory$AbstractCellReadValue and org.knime.chem.types.MolValue are in unnamed module of loader org.eclipse.osgi.internal.loader.EquinoxClassLoader @437d7402)
at org.knime.chem.types.MolCellValueFactory.getValueAsString(MolCellValueFactory.java:1)
at org.knime.chem.types.AbstractStringBasedValueFactory$AbstractCellWriteValue.setValue(AbstractStringBasedValueFactory.java:82)
at org.knime.core.data.v2.WriteAccessRowWrite.setFrom(WriteAccessRowWrite.java:99)
at org.knime.core.data.v2.WriteAccessRowWrite.setFrom(WriteAccessRowWrite.java:1)
at org.knime.core.data.columnar.table.ColumnarRowWriteCursor.commit(ColumnarRowWriteCursor.java:103)
at org.knime.python3.arrow.PythonArrowDataSourceFactory.copyTable(PythonArrowDataSourceFactory.java:194)
at org.knime.python3.arrow.PythonArrowDataSourceFactory.copyTableToArrowStore(PythonArrowDataSourceFactory.java:179)
at org.knime.python3.arrow.PythonArrowDataSourceFactory.extractStoreCopyTableIfNecessary(PythonArrowDataSourceFactory.java:170)
at org.knime.python3.arrow.PythonArrowDataSourceFactory.createSource(PythonArrowDataSourceFactory.java:120)
at org.knime.python3.arrow.PythonArrowTableConverter.createSource(PythonArrowTableConverter.java:107)
at org.knime.python3.nodes.ports.PythonPortObjects$PythonTablePortObject.getDataSource(PythonPortObjects.java:229)
at jdk.internal.reflect.GeneratedMethodAccessor256.invoke(Unknown Source)
at java.base/jdk.internal.reflect.DelegatingMethodAccessorImpl.invoke(Unknown Source)
at java.base/java.lang.reflect.Method.invoke(Unknown Source)
at py4j.reflection.MethodInvoker.invoke(MethodInvoker.java:244)
at py4j.reflection.ReflectionEngine.invoke(ReflectionEngine.java:357)
at py4j.Gateway.invoke(Gateway.java:282)
at py4j.commands.AbstractCommand.invokeMethod(AbstractCommand.java:132)
at py4j.commands.CallCommand.execute(CallCommand.java:79)
at py4j.ClientServerConnection.sendCommand(ClientServerConnection.java:244)
at py4j.CallbackClient.sendCommand(CallbackClient.java:384)
at py4j.CallbackClient.sendCommand(CallbackClient.java:356)
at py4j.reflection.PythonProxyHandler.invoke(PythonProxyHandler.java:106)
at jdk.proxy6/jdk.proxy6.$Proxy78.execute(Unknown Source)
at org.knime.python3.nodes.CloseablePythonNodeProxy.execute(CloseablePythonNodeProxy.java:566)
at org.knime.python3.nodes.DelegatingNodeModel.lambda$4(DelegatingNodeModel.java:180)
at org.knime.python3.nodes.DelegatingNodeModel.runWithProxy(DelegatingNodeModel.java:237)
at org.knime.python3.nodes.DelegatingNodeModel.execute(DelegatingNodeModel.java:178)
at org.knime.core.node.NodeModel.executeModel(NodeModel.java:596)
at org.knime.core.node.Node.invokeFullyNodeModelExecute(Node.java:1284)
at org.knime.core.node.Node.execute(Node.java:1049)
at org.knime.core.node.workflow.NativeNodeContainer.performExecuteNode(NativeNodeContainer.java:603)
at org.knime.core.node.exec.LocalNodeExecutionJob.mainExecute(LocalNodeExecutionJob.java:98)
at org.knime.core.node.workflow.NodeExecutionJob.internalRun(NodeExecutionJob.java:198)
at org.knime.core.node.workflow.NodeExecutionJob.run(NodeExecutionJob.java:117)
at org.knime.core.util.ThreadUtils$RunnableWithContextImpl.runWithContext(ThreadUtils.java:369)
at org.knime.core.util.ThreadUtils$RunnableWithContext.run(ThreadUtils.java:223)
at java.base/java.util.concurrent.Executors$RunnableAdapter.call(Unknown Source)
at java.base/java.util.concurrent.FutureTask.run(Unknown Source)
at org.knime.core.util.ThreadPool$MyFuture.run(ThreadPool.java:123)
at org.knime.core.util.ThreadPool$Worker.run(ThreadPool.java:246)
This only happens in the “Molecule properties” node when run like so:
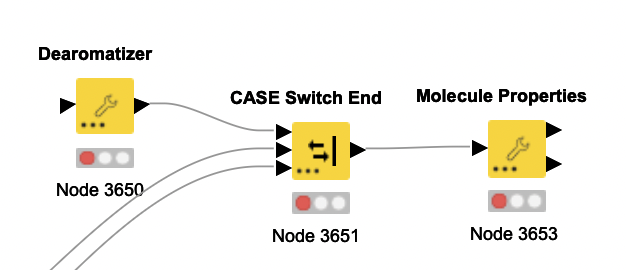
If the two custom nodes are connected directly, there is no problem. I’m using the following code to cast the desired column to Molfile in the “Dearomatizer”:
def execute(self, exec_context: knext.ExecutionContext, input_table: knext.Table):
LOGGER.warning(input_table.schema[self.selected_col].get().ktype.logical_type)
# Load the input table
df: pd.DataFrame = input_table.to_pandas()
int_cols = [col.name for col in input_table.schema if col.ktype == schema.int32()]
# Initialize Indigo
indigo = Indigo()
# Define a function to dearomatize a molecule
def dearomatize_molecule(mol_block):
try:
mol = indigo.loadMolecule(mol_block)
mol.dearomatize()
return mol.molfile()
except Exception as e:
return None # Return None if dearomatization fails
df[self.output_col] = df[self.selected_col].apply(dearomatize_molecule)
LOGGER.warning(dir(cet))
LOGGER.warning(help(cet.MolValueFactory.encode))
df[self.output_col] = df[self.output_col].apply(lambda x: cet.MolValue(cet.MolValueFactory().encode(x)) if x is not None else None)
# force int32 columns to be nullable
df[int_cols] = df[int_cols].astype("Int32")
# Output the modified table
return knext.Table.from_pandas(df)