Hi,
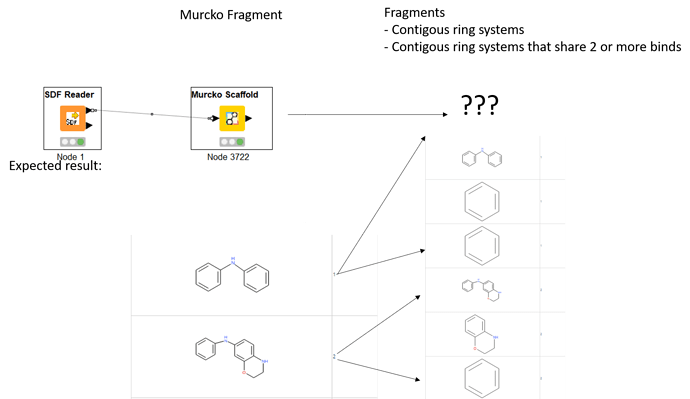
I would like to create a Workflow that generates Scaffolds from an SDF input file.
The steps would be:
- Murcko fragments
- Further fragmentation generating the following scaffold types
- Contiguous ring systems
- Contiguous ring systems that share two or more bonds (bridges between rings)
With the Indigo “Murcko Scaffold” node, I was able to generate Murcko scaffolds with the expected outcome but so far I was not able to reproduce further ring system fragmentation with the available fragmentation nodes (e.g. Vernalis) that resulted in the expected outcome.
I attached an image with molecules expected. It would be nice if someone has an idea on implementing this in KNIME. I also will try if I can achieve this with RDKit + Python but have not done this in the past so if someone can give me a hint using the RDKit tool would also be very nice.
Many thanks in advance.
Best