I have huge CSV files zipped into GZIP to work with.
I know 1 slow way to read them:
- Node “Decompress Files” that wants to write the big CSV file to the HDD first
- “CSV Reader”…
Is it possible to skip the write to disk step?
I have huge CSV files zipped into GZIP to work with.
I know 1 slow way to read them:
Is it possible to skip the write to disk step?
All reader nodes should be able to directly read Gzipped files if the file name ends with .gz.
Please add a http link to this node “All Reader”. I don’t see it in the app’s node repository.
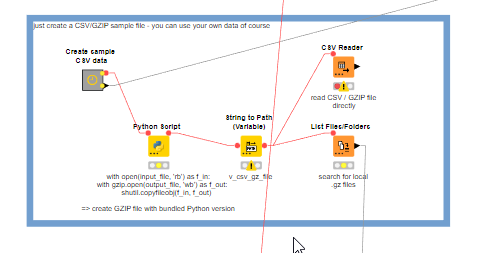
@Iwo you can just use the CSV Reader (CSV Reader – KNIME Community Hub) like this:
A wider approach to import CSV with GZIP an Hive is here:
CSV Reader has a problem with tar.gz file with CSV inside:
ERROR CSV Reader 4:1338 Execute failed: Cannot create a GZIPInputStream directly from a tar archive (C:\iwo_test\13855681Knime_tar_gz.tar.gz).
CSV Reader works with the same not compressed CSV file
@Iwo maybe you can provide us with a sample file without spelling any secrets. There was no mention of TAR before …
@Iwo you can create .tar.gz files and also extract CSV from them like in this example:
import knime.scripting.io as knio
import tarfile
import numpy as np
import pandas as pd
csv_file = knio.flow_variables['File path']
output_file = knio.flow_variables['context.workflow.data-path'] + knio.flow_variables['File name'] + ".tar.gz"
knio.flow_variables['v_tar_gz_file'] = output_file
with tarfile.open(output_file, 'w:gz') as tar:
tar.add(csv_file)
You could give the .tar.gz file as “input_file” and the code would then extract all CSV files into the “output_directory” you have defined. You can adapt these settings:
Last example would extract the first CSV file found in a pandas dataframe and give that back to KNIME. Please note the sample CSV file as pipes has columns separators (|) you might want to adapt that.
kn_example_python_tar_gzip_csv.knwf (76.3 KB)
This topic was automatically closed 90 days after the last reply. New replies are no longer allowed.