Thanks again @Daniel_Weikert  !
!
I very much appreciate it !
… and it motivated me to eventually put in place a possible solution 
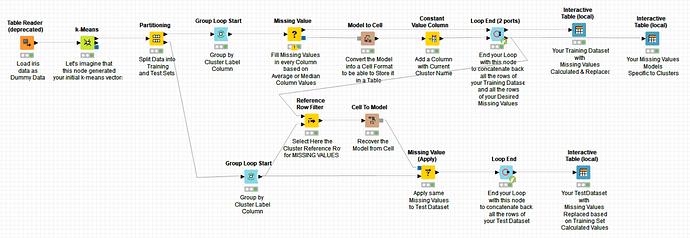
20211201 Pikairos Handling missing value in clustered data.knwf (687.0 KB)
@m1k here you have a workflow which should answer your second question. I did not use a set with missing values as reference but if you replace the data set used here by yours with missing values, it should do the job. Otherwise, let us know and we will amend it.
Hope it helps
Best
Ael
Ps: @Daniel_Weikert yes my background is ML