hi,
i am working with a big dataset of molecules containing the compounds names. Using the proper nodes, KNIME was able to give me the SMILES strings of only 2700 compounds out of 9500 and that’s because for a huge block of compounds the IUPAC name wasn’t reported on the list.
how can I get the IUPAC names from a list of compounds names?
thank you so much in advance!
Margherita
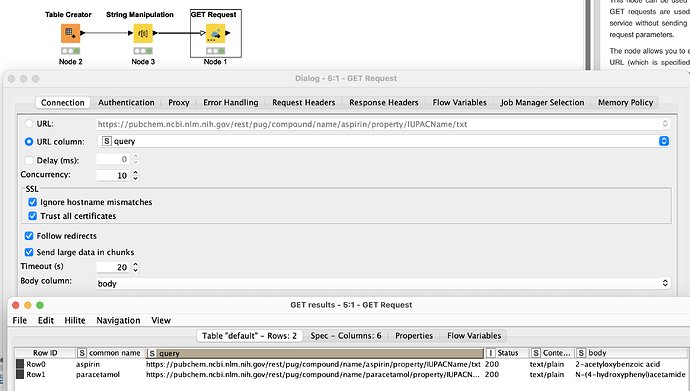
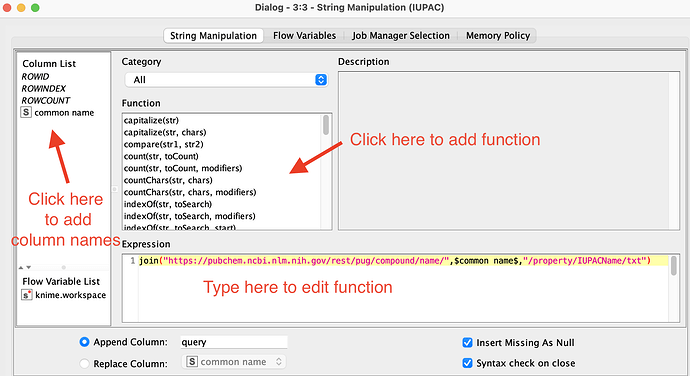
This is where you need to use web-based APIs like PubChem, and where KNIME makes it easy. Try a workflow like the one below, where you use the String Manipulation node to wrap the common_name in a URL like:
join("https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/",$common_name$,"/property/IUPACName/txt")
Change TXT to CSV, JSON or XML if you prefer, then pass the results to the GET Request node to retrieve IUPAC names.
(Psst ![]() . Also try the form https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/aspirin/property/canonicalsmiles/txt for direct SMILES retrieval)
. Also try the form https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/aspirin/property/canonicalsmiles/txt for direct SMILES retrieval)
thank you again for your help!
I hope to figure it out with your instructions
You’re welcome. Let us know how well it works. There are always more tweaks to try!
(the other)
Simon
What do you mean with using the String Manipulation node to wrap the common_name in a URL? I can’t set up the node…
thank you very much you’ve been very kind.
No problem. Let us know if you see any errors. I suspect that you will still have to deal with punctuation in names that are like 2,2’-dichloro-[N-acetyl]somethingane.
Simon
This topic was automatically closed 90 days after the last reply. New replies are no longer allowed.