Hi everyone,
I have a question regarding a specific workflow created by the Knime Community, which can be found and downloaded from the following hyperlink:
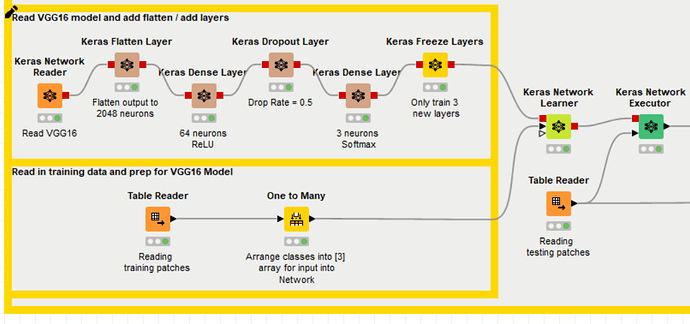
The workflow uses a Keras Transfer Learning to predict Cancer Type from Histopathology Slide Images. Here is a brief description of how the workflow works. The target variable was categorized into three different cancer type classes:
I really appreciated the potential of this workflow, so I am trying to replicate it on a new dataset I found on the web with similar predictive purpose.
The main difference of my dataset is that the target variable I want to analyze is binary, since it only exhibits two realizations: the presence, or the absence, of a pneumothorax in each x-ray image of the dataset.
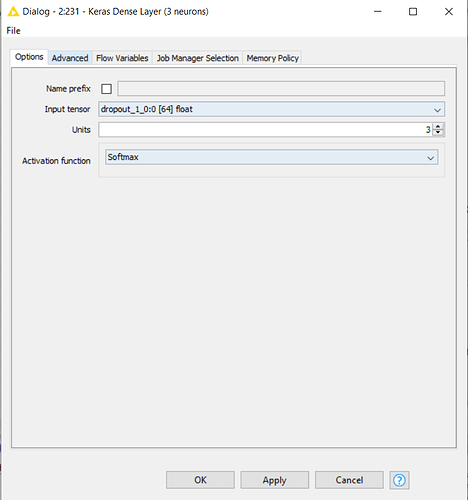
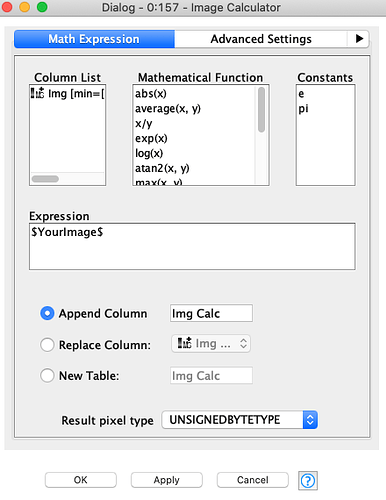
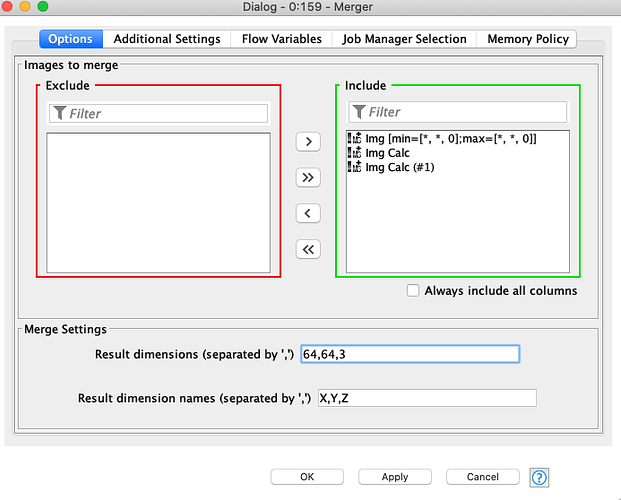
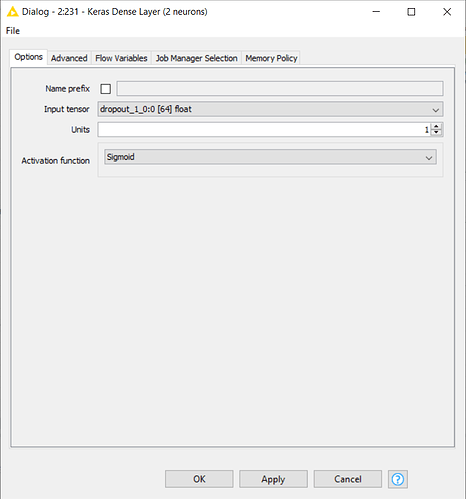
Because of this, in order to replicate the workflow on my dataset, I should first change the numerosity of neurons in the output layer from three neurons to two, and then theorically I could run the pre-trained model on my data. But here’s the problem: I have tried to reconfigure the Keras Dense Layer Node (which you can see in the following uploaded capture) by reducing the Units from 3 to 2 and by changing the Activation Function into a Sigmoid, which is more suitable for a binary target variable.
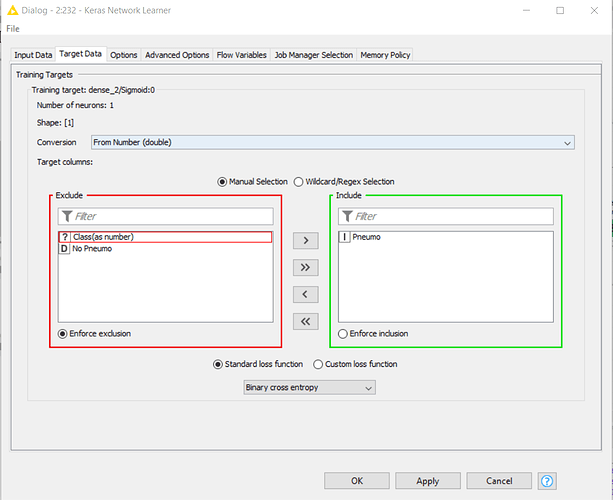
Then, when I run the Keras Network Learner, i get the following log error:
ERROR Keras Network Learner 0:233:232 Execute failed: Node training data size for network input/target ‘input_2:0’ does not match the expected size. Neuron count is 12288, batch size is 64. Thus, expected training data size is 786432. However, node training data size is 262144. Please check the column selection for this input/target and validate the node’s training data.
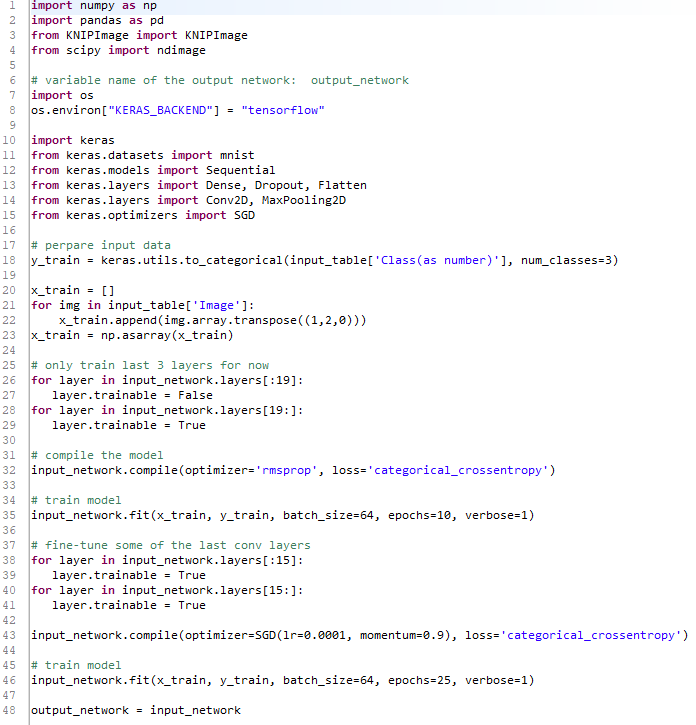
Lastly, I found one more interesting workflow description webpage which breaks each of the previous nodes into a Python code. Unfortunately I am not really good at coding so I could not solve the issue by myself:
https://www.knime.com/blog/transfer-learning-made-easy-with-deep-learning-keras-integration
I hope you can help me on this even if I know it is really complicated, you would save my project!
Thank you all in advance,
Nicola