Friends, I’m happy to share with you my new open-source project – knime2py.
GitHub - vitaly-chibrikov/knime2py: KNIME project to Python workbook converter
If you work in KNIME you know how fast new prototype of DS project can be created in it. And, at the same time, how complex already developed KNIME project can be. But maybe you, as I am, fill the lack of ability to export your KNIME project to .py or .ipynb files.
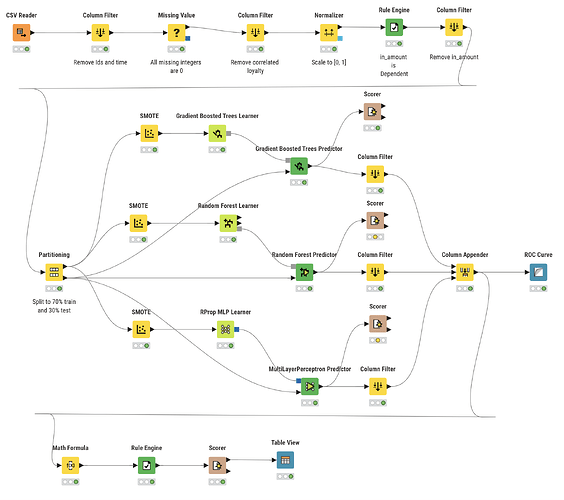
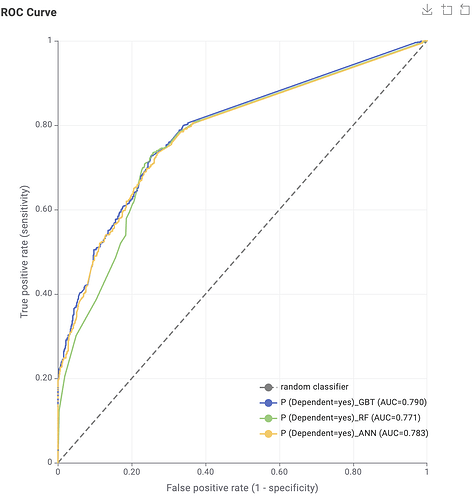
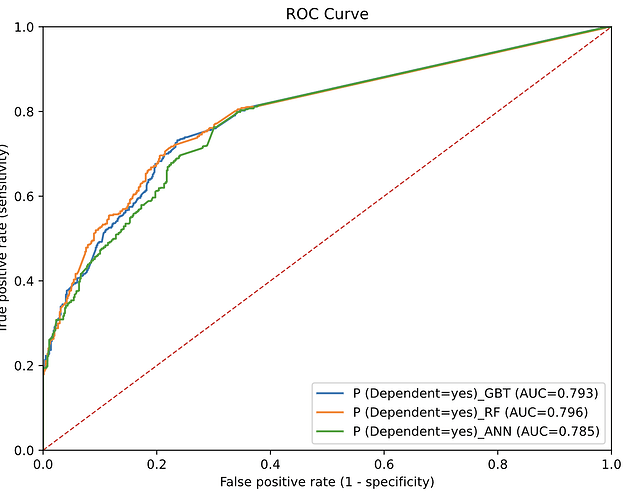
So, I decided to start my new open-source project “knime2py” which is a code-generation and KNIME→Python exporter: it parses a KNIME workflow, reconstructs its nodes and connections, and emits runnable Python “workbooks” (Jupyter notebook or script) by translating supported KNIME nodes into idiomatic pandas / scikit-learn code via a pluggable node registry. Alongside the executable code, it also writes a machine-readable graph (JSON) and a Graphviz DOT file, preserving port wiring and execution order so the generated Python mirrors the original workflow.
The list of already supported nodes can be found here: knime2py — Implemented Nodes
Even if the node you use is not implemented, exporter creates cell for it with initialization of parameters from the node’s settings.xml.
Fill free to try it. I will be happy to discuss any questions.