Hi Guys,
One of the most difficult things for me are the Lookups. I attached the pic to help me explain. I want to replace a value in one file if it found a value in the other file as shown in the figure.
Thanks in advance
Hi Guys,
One of the most difficult things for me are the Lookups. I attached the pic to help me explain. I want to replace a value in one file if it found a value in the other file as shown in the figure.
Thanks in advance
Hi @VAGR_ISK , I think I understood what you need, but I have to say, the naming of columns as Row0, Row1, etc, can be confusing.
Can you please give some sample data to work with? We’d need samples for both files.
Hi @VAGR_ISK,
which final result do you expect.
Do you simple want to count the occurrence for each protein a column?
BR
Hi @morpheus , I think the final result would be a modified File 2, where some 1 have been replaced by 2, based on the rule that @VAGR_ISK explained.
Hi @bruno29a, You are correct when I transpose the table the Columns are named rows. My apologies. I will be super happy if you can help.
Here there is a sample of both files and the expected outcome in a third file. They are organized in different sheets and the Xcel formula is also there.
Thanks in advance,
Example.xlsx (65.7 KB)
Thanks for the sample @VAGR_ISK , and just to confirm, you would have much more columns in the real data file, correct?
Hi Morpheus,
Please follow the white rabbit… 
Yes, I basically want to have 2 in cells of File number 2 that are found in file number 1. I add an example with the excel formula that I use here. I guess that will be easier to understand if you see it.
Thanks for the help.
Example.xlsx (65.7 KB)
Hi @VAGR_ISK , this isn’t a solution to the overall problem, but a suggestion re the column names following the transpose.
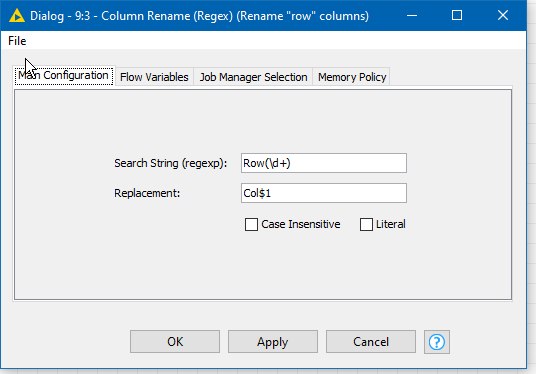
You can put them through a Column Rename (Regex) node to rename all the RowN columns to ColumnN (or anything else you wish)
For example:
So (and the data shown here was typed in by me before you posted your example.xlsx, so it isn’t accurate but is representative)
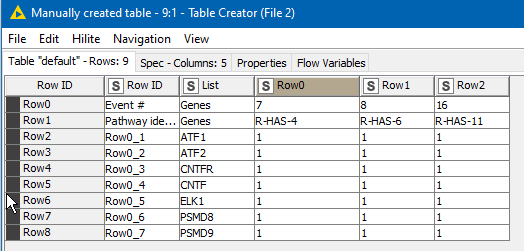
becomes
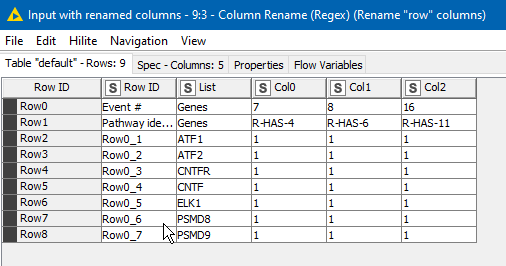
This at least removes one complication from the discussion 
Hi @VAGR_ISK , sorry I had to step out earlier for 2hrs, but I have to say, I was eager to get back to work on this. Finally I’m back, and I put something together using your sample file.
The workflow looks like this:
And the final results looks like this:
I looks the same as the results you provided with your sample - please double check.
You can add some tweaks to get the final result as you like, but the core logic is in this workflow.
You can rename the columns by extracting the column headers from the original file and replace with them in the final results - I tried to do this, but Knime cannot extract header names made of numerical values, it renames them as Column0, Column1, etc…, you can may be rename the columns to something else beforehand.
All in all, the logic that I’ve used is, for each row from File 2, I go check if the List exist in each column of File 1, and if found, then return value “2”, else return value “1” - this is based on the assumption that by default, all values in File 2 are 1. If that is not the case, then this does not really work, or you could split the data into 2 categories, those that defaults to 1, and those that don’t.
Here’s the workflow:
Lookup in different files with CountIf.knwf (93.4 KB)
EDIT: I added a bit of documentations to the workflow:
Here’s the new version of the workflow that contains the documentations:
Lookup in different files with CountIf.knwf (95.7 KB)
As is often the case, there are different approaches, so here I’ve added what I came up with.
Your existing data is in “tabulated format” which is difficult to work with as it is split across multiple columns.
For such a lookup to work, without loops, you require your data in “row” format. This would be achieved by unpivoting both of your tables.
After that you want to create a “key” for each data item which comprises the name of the protein, and the original column name it was in, and count for each original column, the number of times any given protein appears in File 1 which can then be used in a lookup using “Cell Replacer”.
Where an item doesn’t appear, we replace the missing looked-up count with 0. We can add that lookup value to the original column value (in File 2) for that key, then pivot it back into the original tabular format, removing the now-redundant key field and anything else we used for intermediate values, and out comes the result.
I used your original “RowN” naming for the columns initially which I then converted to “ColN”., which is how they are in the output.
OMG Takbb. That is impressive. I have to say, I am sure, I will never come to this solution since I am a beginner using KNIME. If KNIME will have a type of node that could mimic the Excel lookups like this, I will definitively move 100% KNIME and leave excel, which is good but very slow sometimes. Also, the problem with these complex multistep processes is that one has no way to be sure that a small mistake changed the whole result. At least with the Countif function in Excel, you can visually carefully compare it, understand and repair the problem.
Cool @bruno29a. Thanks a lot. That is impressive. I have to say, I am sure, I will never come to this solution since I am a beginner using KNIME. If KNIME will have a type of node that could mimic the Excel lookups like this, I will definitively move 100% KNIME and leave excel, which is good but very slow sometimes. Also, the problem with these complex multistep processes is that one has no way to be sure that a small mistake changed the whole result since one can not check Big data cell by cell. Ideas?. At least with the Countif function in Excel, you can visually carefully compare it, understand and repair if there is a problem.
The number 1 that I put in the FIle 2 is just to avoid an additional step of putting one in proteins that are not found in File 1, but one can start with no numbers in the cells of FIle 1. But, yes if… always the file 1 will have 1 only.
Thanks again. This lookup problem is a bit trouble some, but I have to say I will surely come with some other problems that will brake your mind…
Cheers.
It might be… I actually got it from a paper somewhere years ago.
Hi @VAGR_ISK,
the white rabbit was shoot by the hunter… 
you can solve it also with simple check of the occurency of a protein and join it to the reference list. The complexity of the workflow is mainly influenced by the final output.
Example.knwf (287.6 KB)
BR
Hi @bruno29a, just to let you know the solution did work well for a shorter number of columns. However, when I placed your proposed pipeline into a bigger list of proteins, I have to stop it because the looping took a lot of time.
This topic was automatically closed 7 days after the last reply. New replies are no longer allowed.