Dear knimers,
Imagine you have to import a bunch of files (say 100) into your knime workflow. The file format contains 10000 floating point numbers (doubles). Unfortunately, the files are in some weird proprietary file format. Fortunately, for almost every proprietary file format someone made a Python library that understands the file format. So the obvious solution is to write a small Python script that reads the file and returns a Pandas dataframe that Knime understands. But we are not dealing with a single file, we are dealing with whole bunch of files. Question is, should we loop over those files using a Knime loop or should we put the loop in the Python script?
Let’s simulate this situation by generating random numbers inside a Python script. To keep the workflow scalable, the information for every new iteration (file) should be put in a new table row, not in a new column. And, instead of putting the values of one iteration in 10000 columns, we put all 10000 values in a single list cell.
The Python script without the loop then looks like:
import numpy as np
n = 10000
output_table = input_table.copy()
z = np.random.randn(1,n).tolist()
output_table = output_table.assign(Out=z)
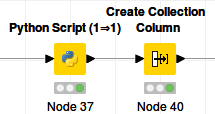
The corresponding Knime workflow looks like:
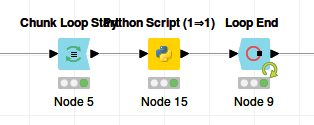
We are simulating loading one file per iteration, so the chunk size is 1.
If we include the loop in the Python script, the script becomes:
import numpy as np
n = 10000
output_table = input_table.copy()
z = np.zeros((len(input_table),n)).tolist()
output_table = output_table.assign(Out=z)
for i, row in output_table.iterrows():
row['Out'] = np.random.randn(1,n).tolist()[0]
output_table.loc[i] = row
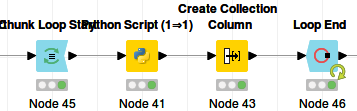
In this case the Knime workflow is only the Python script:
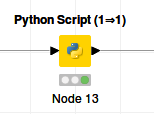
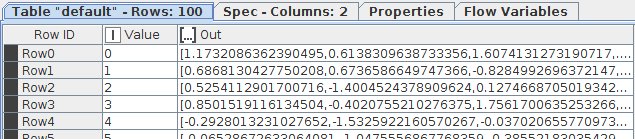
In both cases the output looks like:
The first method takes 173 seconds.
The second methods takes 4 seconds.
How about 100 x 100000 values?
In this case the first method takes option takes 198 seconds.
The second methods takes 26 seconds.
Conclusion: if workflow execution speed matters, it is absolutely worthwhile to write the Python script in such a way that it includes the loop. It is obvious that launching and exiting the Python environment, and serializing and deserializing the data takes so much time that this should be limited as much as possible.
It should be noted that because Python/numpy/pandas does everything in memory, one may run out of memory for large datasets. In this case it would be wise to have both a loop in the Python code and a Knime Chunk Loop with a reasonably large but not too large chunk size.
The workflow: KNIME_project2.knwf (27.2 KB)
 I think this is the case as long as Python has memory available. I would love to see an example where the opposite is true…
I think this is the case as long as Python has memory available. I would love to see an example where the opposite is true…