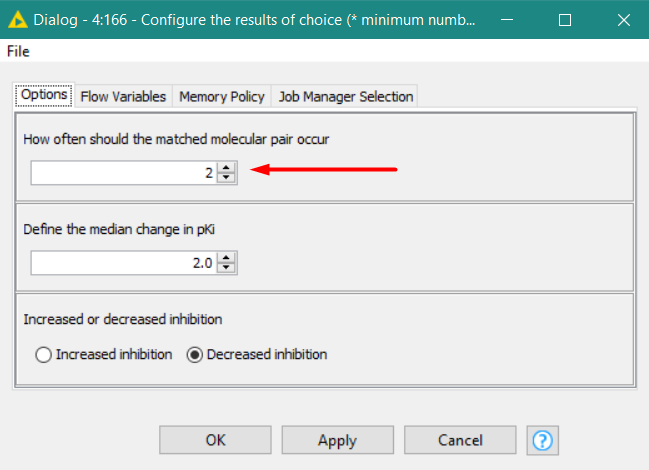
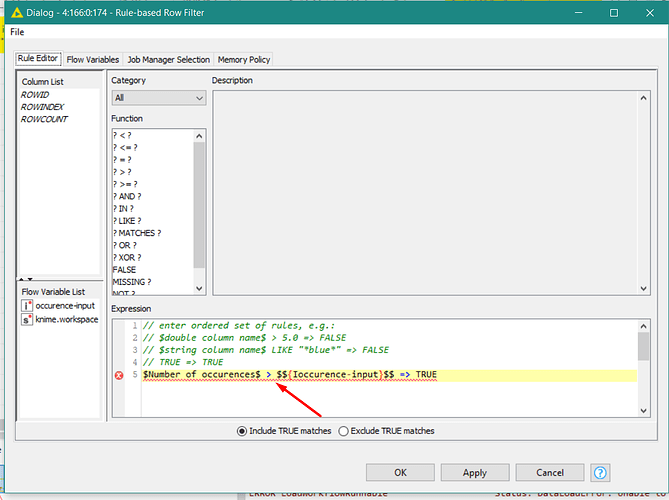
This workflow demonstrates how to to matched molecular pair analysis in KNIME and proposes a way how to deal with biological assay data that includes a qualifer. The data has been retrieved from ChEMBL27 and contains Ki values of hERG. The human ether-à-go-go-related gene (hERG) product is a potassium channel whose inhibition can cause potential lethal cardiac arrythmia, so introducing transformations to drug candidates that reduce hERG inhibition is desired. As MMP analysis results in a range of values, we will take the median to select transformations to investigate further. The Interactive View contains a table with all transformations that match the criteria defined by the user. When a transformation is selected, the details show up in a second table underneath.
This is a companion discussion topic for the original entry at https://hub.knime.com/-/spaces/-/latest/~brARDPUWDy9VG8CM/