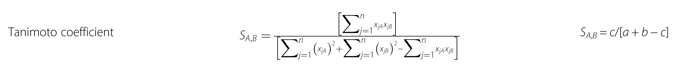Dear KNIMErs,
The Bit Vector Distances node contains an option (i.e. “Tversky distance (Tanimoto/Dice)”) with which, in combination with the Similarity Search node, one can perform a similarity search based on the Tanimoto coefficient. This type of search is especially important in the cheminformatics context.
Nevertheless this option takes into account only the case where you have dichotomous variables (i.e. a bit vector). Another scenario where the possibility to calculate the Tanimoto distance is also very important is the case where you have continuous variables or count-based fingerprints (i.e. a byte vector). For this reason I think it would be very beneficial for the KNIME community to include the possibility of calculating the Tanimoto coefficient for byte vector. Would this be possible?
Here is the formula:
Here, S denotes similarities, xjA means the j-th feature of molecule A . a is the number of on bits in molecule A , b is number of on bits in molecule B , while c is the number of bits that are on in both molecules. On the left part of the figure there is the formula for continuous variables, while on the right part, the formula for dichotomous variables.
The formula has been defined, inter alia, in the following scientific publications:
- Willett P. J. Chem. Inf. Comput. Sci. 1998, 38, 983-996
- Bajusz D. et al. Journal of Cheminformatics (2015) 7:20
Thanks in advance for your answers!