KNIME 4.7.3
Mac/Lin/Win
I have some cdx files in a table as binary objects. When I try to pass the table to the OpenBabel node, that column doesn’t appear in the list of selectable columns – only string type columns appear.
If I use the Binary Objects to Strings node before passing the table to the OpenBabel node, I can select the column, select the input format as “cdx – ChemDraw binary format [Read-only]”, and SMILES as the output format, but the conversion fails silently. No error messages in the console, only missing “?” data in the new appended column.
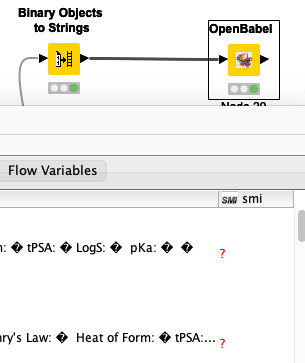
I have tried using all of the Decoding formats in the Binary Objects to Strings node (UTF-8, US-ASCII, etc), and both bundled (Lin/Win) and manually installed (Mac/Lin/Win) versions of the babel executable. No combination is successful.
I can write the binary data to a file (in a loop) using the Binary Objects to Files node, then use the External Tool node to run the babel executable, and that works OK - I get SMILES strings, mol files, etc as output.
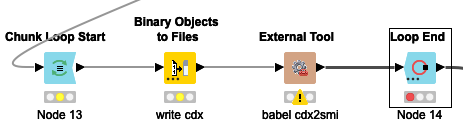
But that means that I need to install the babel executable on any system that wants to run the workflow.
Does anybody know how to pass cdx data to the OpenBabel node so that I can use the bundled version of babel in the OpenBabel node?
(the other)
Simon
Hi the other Simon 
thanks for reporting this, together with a workaround. We’d be happy to take a look at this.
By any chance, do you have a small example workflow ready to share for any non-moleculists wanting to give this a shot?
Kind regards
Marvin
1 Like
OK Marvin, attached a very simple workflow with example data. I don’t have access to the original tool anymore, so had to use another tool to generate the data. So there may be differences, but it seems to duplicate the original problem.
(the other)
Simon
openbabel-binary-example.knwf (32.0 KB)
1 Like
Hi @marvin_kickuth,
Any updates available on this?
Thanks,
(the other)
Simon
Hi @sauberns,
Thank you for reporting and providing the example workflow.
We have created a ticket for this (AP-20793).
Continuing the discussion from OpenBabel binary inputs?:
Hi @marvin.kickuth, @armingrudd,
With the release of 5.3, do we have any view on a solution for ticket (AP-20793)?
Thanks,
(the other)
Simon
Hi @sauberns,
Unfortunately, there is no update regarding this ticket yet.
I moved the topic to Feedback & ideas so it will receive an update as soon as the ticket is done.
Thank you for your patience! 
Hi @armingrudd - Any update on this? I’m trying to read .cdx files and use the same process with openbabel but it just runs and runs - nothing happens.
My workflow looks a little like this -
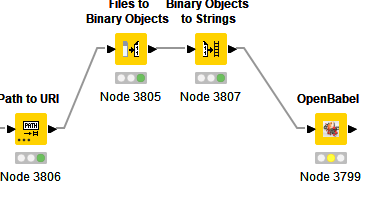
I’ve tried not doing the files to binary objects but that doesn’t work either.
Thanks,
Angie
Hello @chemgirl36,
Unfortuantely, not yet and thanks for your patience. I just shared this with our devs again.
1 Like
It seems openbabel is also having an issue with converting cdxml to smiles or anything usable - I’m not sure why it isn’t working properly.


