Hello, colleagues,
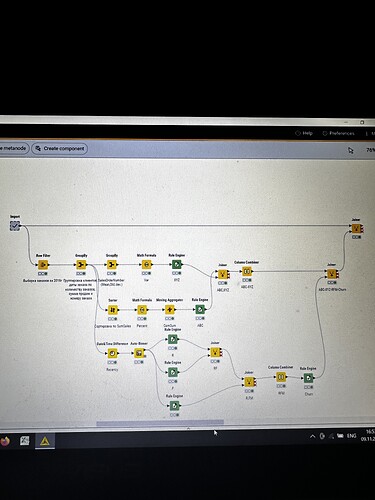
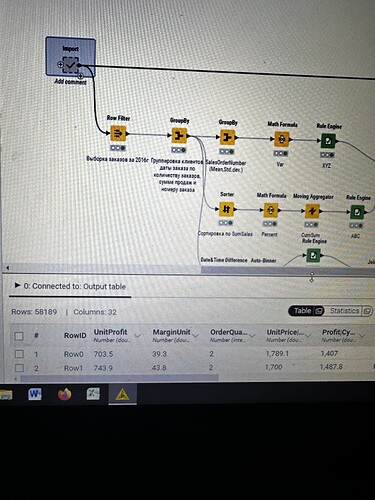
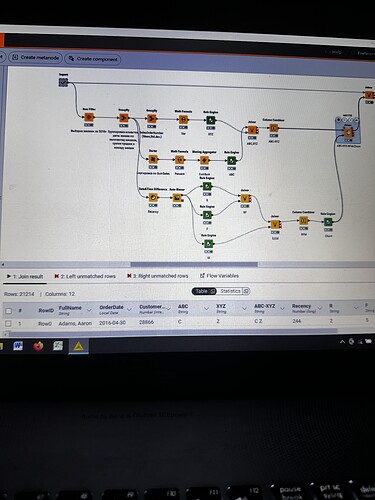
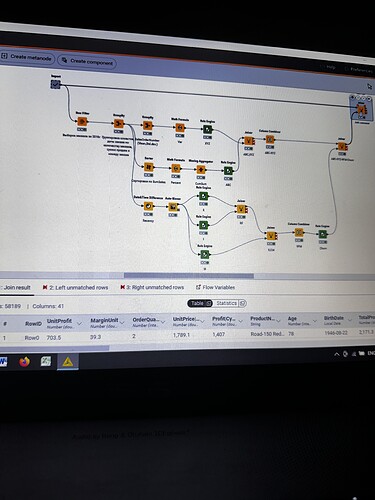
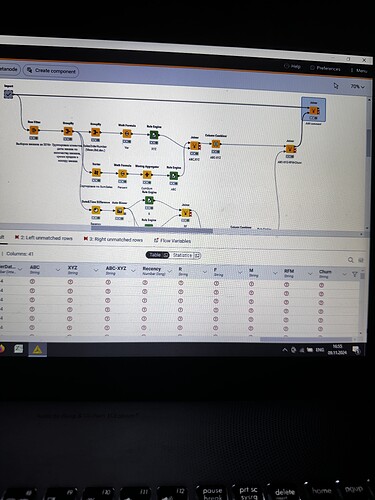
I have strange situation with my workflow. I executed import metanode (58.189 rows and 32 columns), then I performed the ABC-XYZ-RFM-Churn analysis (at the final homer node there are 21.214 rows and 12 columns). Then I tried to join two parts: import metanode and the last joiner of ABC-XYZ-RFM-Churn analysis, using the left outer join with the keys Customer Key and Order date (if I use only customer key, I have increased quantity of rows), and include all columns from import metanode (32 columns: unit profit, margin unit, order quantity, unit price, profit, age, birthdate, total product cost, sales amount, full name, gender, yearly income, education, occupation, date first purchase, customer city, customer country, order date, sales order number, subcategory, category, order date (год+квартал), order date (год+месяц), order date (год+неделя), customer key, product key, commute distance, product line, order date (год, количество), order date (год, число), order date (год, строка), product name) and from ABC-XYZ-RFM-Churn analysis last joiner only new generated columns (ABC, XYZ, ABC-XYZ, Recency, R, F, M, RFM, Churn), which are not inside the Import metanode. Then I put the flag in Append custom suffix if there are duplicate column name. Then put the flag in Create new in case of RowIDs and push OK. Then I have 58.189 rows and 41 column with missing values in ABC, XYZ, ABC-XYZ, Recency, R, F, M, RFM, Churn. I changed the matching concept and checked the quantity of rows and columns at all joining that I have before, nothing changed…I have no idea, HELP!