Dear KNIMEler,
I’m observing some problems - from time to time - with GroupBy nodes being executed within a Parallel Chunk Loop. Although having set the ‘Maximum unique values per group’ to a very high (absurd) number (100.000.000) I get: Maximum unique values number too big when doing a group-by on a table with ~50.000 columns, calculating some statistics (mean, median, mad, std.dev) for all records for just one DOUBLE column. As this error is only happening from time to time (thus hard to re-construct) and only within parallel execution I assume some memory problems.
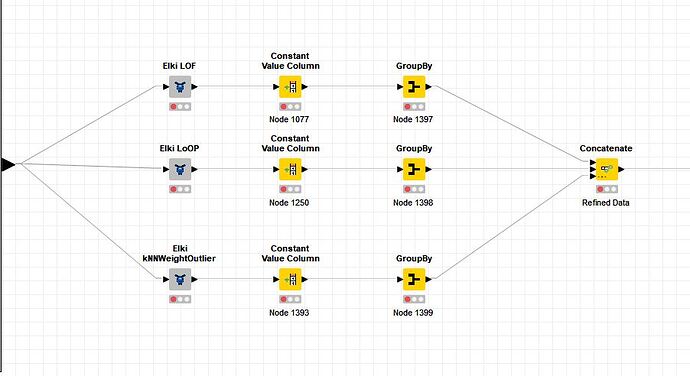
The Workflow
The WF shown on the screenshot attached (see image) calculates for one table - e.g. having 50.000 rows - three outlier scores via the 3 components named Elki. The GroupBy nodes are set-up to calculate the median, mean, MAD and stddev for each outlier-score (using all rows, setting the ‘Maximum unique values per group’ to the absurd high value 100.00.000). All three GroupBy modes have the same settings.
The WF shown is executed with a counting Loop (doing Parameter shuffling) which is embedded in a Parallel Chunk Loop with 3 chunks defined. That means, that 3 instances of the shown WF are running in parallel.
The Problem
When running my WF it can/might happen, that the ‘Parallel Chunk End’ loop reports an execution error (after 30% completion) in one of the parallel instances! Inspecting this error shows the above mentioned problem of the GroupBy node - see log attached! The strange part is:
- The table fed into the node has about 50k rows
- Simply restarting the GroupBy node manually works
Furthermore,
- outside a Parallel Chunk-loop I never encountered any problems with GroupBy nodes.
- it happens on Linux and on Windows
- restarting KNIME seems to bring a delay
- I have seen/noticed this problem over several KNIME versions (4.2 - 4.5)
Any idea?
Erich
log.txt (7.0 KB)