Hi,
I was trying to use a Python 1=>2 Scripting node to transform Molecules and then output them into two tables.
KNIME crashed multiple times trying to display the table, always non reproducible showing me either the desired table or just weard numbers, but each time with a pointer error.
I tried to go back to a very easy script and found a (or the?) problem:
Firstly if I create the two tables separately with eg.
output_table_1 = pd.Dataframe({“a” : [molecule,molecule], “b”:[1,2]})
output_table_2 = pd.Dataframe({“a” : [molecule,molecule], “b”:[1,2]})
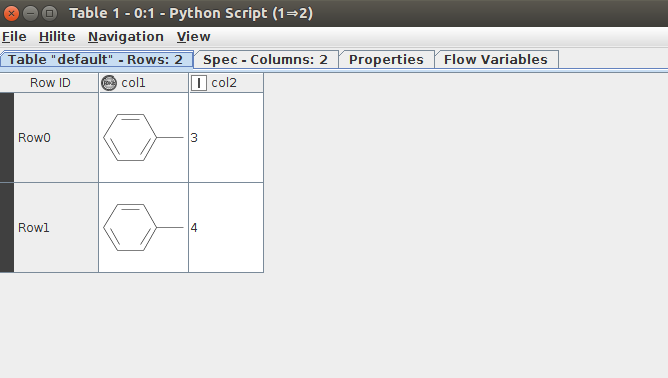
I see the two tables with the RDKit Molecule and everything works fine.
If I do something like
x = pd.Dataframe({"a" : [molecule,molecule], "b":[1,2]})
output_table_1 = x
output_table_2 = x
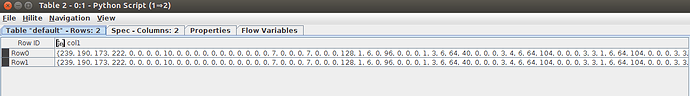
The first table is displayed correctly but the second one shows me weard numbers instead of the molecule.
This happens with the apache arrow as well as the flatbuffer serialization.
I have attached parts of the error messages (I can attach the whole if needed) and a simple workflow to reproduce the error.
I am not sure if I am doing something weard or if there is somehow a communication problem between rdkit, python and KNIME. (It works without the RDKit Molecules)
System: Ubuntu 16.04 LTS
KNIME: 3.7.1
RDKit KNIME integration 3.6.0.v201903281548
Python: Anaconda environment with python 3.6.7 and rdkit 2019.03.1.0 (error was also there for the latest 2018 release, just upgraded today) pyarrow 0.11.0
I am happy to provide more information if needed.
Thanks in advance for your ideas!
jennifer
Python_fail.knwf (22.3 KB)
errs.txt (4.4 KB)