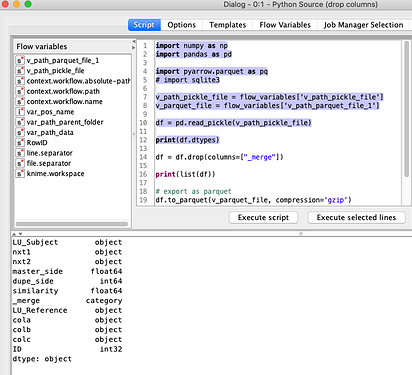
Well, changing the datatype of the object columns to string does not seem to work in pandas. Apparently, object is what is used for strings.
After executing my module, called from a “Python Source” node, the dataframe generated, per .dtypes, is as follows:
LU_Subject object
nxt1 object
nxt2 object
master_side float64
dupe_side int64
similarity float64
_merge category
LU_Reference object
cola object
colb object
colc object
ID int32
I obtained the above by executing “print(my_dataframe.dtypes)” within my source module, that is called from “Python Source” node, via the following code in the “Python Source” node:
import sys
sys.path.append(’/Homebase/Source Code/PycharmProjects/Fuzzy5’)
import main_fuzzy_match_v1 as fm
output_table = fm.fnFuzzyMatch(‘Silver’,
‘Test’, ‘SELECT [LU], [nxt1], [nxt2] FROM [SUBJECT_test3]’, ‘LU’,
‘Test’, ‘SELECT [LU], [cola], [colb], [colc] FROM [REFERENCE_test3]’, ‘LU’,
‘SG_Stage_1_Simpledata’)
Values in the “_merge” (category) column (obtained by printing to stdout from my source module) are as follows:
0 both
1 both
2 both
3 both
4 both
5 both
6 both
7 left_only
8 both
9 left_only
The error in the KNIME console is:
ERROR Python Source 0:5 Execute failed: fill value must be in categories
What appears to be the relevant portion of the log-file, is:
2020-10-06 17:57:15,585 : ERROR : KNIME-Worker-14-Python Source 0:5 : : Node : Python Source : 0:5 : Execute failed: fill value must be in categories
org.knime.python2.kernel.PythonIOException: fill value must be in categories
at org.knime.python2.util.PythonUtils$Misc.executeCancelable(PythonUtils.java:297)
at org.knime.python2.kernel.PythonKernel.waitForFutureCancelable(PythonKernel.java:1682)
at org.knime.python2.kernel.PythonKernel.getDataTable(PythonKernel.java:993)
at org.knime.python2.nodes.source.PythonSourceNodeModel.execute(PythonSourceNodeModel.java:96)
at org.knime.core.node.NodeModel.execute(NodeModel.java:747)
at org.knime.core.node.NodeModel.executeModel(NodeModel.java:576)
at org.knime.core.node.Node.invokeFullyNodeModelExecute(Node.java:1236)
at org.knime.core.node.Node.execute(Node.java:1016)
at org.knime.core.node.workflow.NativeNodeContainer.performExecuteNode(NativeNodeContainer.java:558)
at org.knime.core.node.exec.LocalNodeExecutionJob.mainExecute(LocalNodeExecutionJob.java:95)
at org.knime.core.node.workflow.NodeExecutionJob.internalRun(NodeExecutionJob.java:201)
at org.knime.core.node.workflow.NodeExecutionJob.run(NodeExecutionJob.java:117)
at org.knime.core.util.ThreadUtils$RunnableWithContextImpl.runWithContext(ThreadUtils.java:334)
at org.knime.core.util.ThreadUtils$RunnableWithContext.run(ThreadUtils.java:210)
at java.util.concurrent.Executors$RunnableAdapter.call(Executors.java:511)
at java.util.concurrent.FutureTask.run(FutureTask.java:266)
at org.knime.core.util.ThreadPool$MyFuture.run(ThreadPool.java:123)
at org.knime.core.util.ThreadPool$Worker.run(ThreadPool.java:246)
Caused by: org.knime.python2.kernel.PythonIOException: fill value must be in categories
at org.knime.python2.kernel.messaging.AbstractTaskHandler.handleFailureMessage(AbstractTaskHandler.java:146)
at org.knime.python2.kernel.messaging.AbstractTaskHandler.handle(AbstractTaskHandler.java:92)
at org.knime.python2.kernel.messaging.DefaultTaskFactory$DefaultTask.runInternal(DefaultTaskFactory.java:256)
at java.util.concurrent.FutureTask.run(FutureTask.java:266)
at java.util.concurrent.Executors$RunnableAdapter.call(Executors.java:511)
at java.util.concurrent.FutureTask.run(FutureTask.java:266)
at java.util.concurrent.ThreadPoolExecutor.runWorker(ThreadPoolExecutor.java:1149)
at java.util.concurrent.ThreadPoolExecutor$Worker.run(ThreadPoolExecutor.java:624)
at java.lang.Thread.run(Thread.java:748)
Caused by: Traceback (most recent call last):
File “C:\Program Files\KNIME\plugins\org.knime.python2_4.2.2.v202009241055\py\messaging\RequestHandlers.py”, line 96, in _handle_custom_message
response = self._respond(message, response_message_id, workspace)
File “C:\Program Files\KNIME\plugins\org.knime.python2_4.2.2.v202009241055\py\messaging\RequestHandlers.py”, line 218, in _respond
data_bytes = workspace.serializer.data_frame_to_bytes(data_frame_chunk, start)
File “C:\Program Files\KNIME\plugins\org.knime.python2_4.2.2.v202009241055\py\Serializer.py”, line 205, in data_frame_to_bytes
data_bytes = self._serialization_library.table_to_bytes(table)
File “C:\Program Files\KNIME\plugins\org.knime.python2.serde.flatbuffers_4.2.0.v202006261130\py\Flatbuffers.py”, line 685, in table_to_bytes
col.fillna(value=’’, inplace=True)
File “C:\Users\paper\anaconda3\envs\py3_knime_auto\lib\site-packages\pandas\core\series.py”, line 3425, in fillna
**kwargs)
File “C:\Users\paper\anaconda3\envs\py3_knime_auto\lib\site-packages\pandas\core\generic.py”, line 5408, in fillna
downcast=downcast)
File “C:\Users\paper\anaconda3\envs\py3_knime_auto\lib\site-packages\pandas\core\internals.py”, line 3708, in fillna
return self.apply(‘fillna’, **kwargs)
File “C:\Users\paper\anaconda3\envs\py3_knime_auto\lib\site-packages\pandas\core\internals.py”, line 3581, in apply
applied = getattr(b, f)(**kwargs)
File “C:\Users\paper\anaconda3\envs\py3_knime_auto\lib\site-packages\pandas\core\internals.py”, line 2006, in fillna
values = values.fillna(value=value, limit=limit)
File “C:\Users\paper\anaconda3\envs\py3_knime_auto\lib\site-packages\pandas\util_decorators.py”, line 178, in wrapper
return func(*args, **kwargs)
File “C:\Users\paper\anaconda3\envs\py3_knime_auto\lib\site-packages\pandas\core\arrays\categorical.py”, line 1756, in fillna
raise ValueError(“fill value must be in categories”)
ValueError: fill value must be in categories