I’m having problems with my R Snippets. In particular, some nodes are not receiving the expected “knime.in” from the upstream node (in this case joiner). This causes an attempt to execute the node to progress to 15% and then die with an error of
Execute failed: R evaluation failed.: “bt”
Strangely, the “column list” on the left side of the setup window for the snippet shows valid values but the “workspace” on the right hand side is empty.
This is KNIME 3.5.2 in 64bit Windows. I just checked for updates in KNIME while writing this and none were reported as available. This R Snippet worked yesterday. R scipting extension shows version 3.1.103v201607041324
I can’t explain why this is happening the way that it is. Some times R Snippet will receive knime.in as expected, and then other times on the right hand side instead I see
name (type)
bt (list)
colCOunt (integer)
cols (list)
i (integer)
knime.col.names (character)
knime.row.names (character)
rowCount (integer)
It seems that some tables will never produce knime.in while some will do it erratically. I have now been able to reproduce this error in KNIME 3.5.2 in 64 bit Linux as well, even when I allocate 12GB RAM for the heap.
Can you share the workflow that reproduces the issue?
Marten
I can share the workflow, but the original input file is ~300MB and I cannot share it currently. Essentially what I’m finding though is that after using the Joiner node - which produces reasonable output when I view the output of said node - the R Snippet instead of receiving the table from Joiner receives the odd items mentioned previously. This makes the R Snippet fail and the rest of my workflow cannot progress. I am having this problem now both in Windows and Linux with KNIME 3.5.2
KNIME_smallmol_troubleshooting.knwf (37.1 KB)
This workflow shows the behavior I’ve been seeing. The file for “Input File” is a mzML file that is then read by FileToTable (which comes from the OpenMS tool set). Everything is fine up through the Joiner, but then things fall off the rails when we get to the R Snippet. The R Snippet node usually runs for a few seconds - showing progress up to 15% - before it fails and returns the problems I’ve reported here.
I have found that for what I was doing in R - getting the previous value of a row into the current row as a new element - I can accomplish the same using the “Lag Column” node. This is fine for what I’m doing here but it doesn’t resolve the oddity of the R Snippet not getting the table from KNIME as it ought to. Furthermore the table that KNIME isn’t passing correctly on to R Snippet (from Joiner) is correctly going to Lag Column without any problem.
Hi lparsons42!
I cannot run the attached workflow, since it’s missing the input file, so I can only guess:
You may want to try increasing the “Rserve receiving buffer size limit” to something larger than 256 or even set it to 0 for unlimited. You will find this option underneath “Preferences > KNIME > R” in the KNIME Analytics Platform.
I hope this solves your problem.
Cheers, Jonathan.
Has anyone solved this?
I’m seeing this error with a 10k x 10k table of doubles (~900 Mb). I have xmx set to 32G and the R buffer set to 0. (unlimited). Also tried adding cache node prior to the R-Snippet.
Using Knime 3.6.0; Win 10 64-bit; on PC with 64G RAM.
Does anyone know the size limit for knime tables -> R?
Hi @SOH979!
You should try a smaller batch size, otherwise too much data is being transferred at once, which causes Rserve to fail:
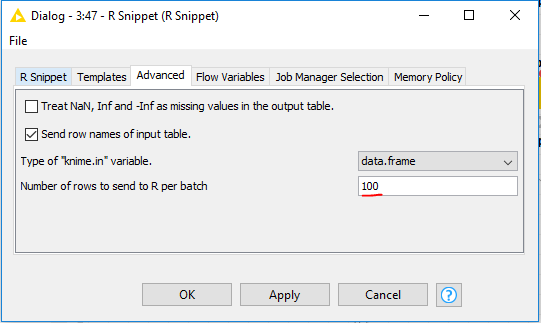
Hope that solves your problem!
Cheers, Jonathan.
PS: Note there is a tradeoff, each batch has a small overhead, hence less batches is usually faster, so try keeping the number as high as possible and small as necessary.
This topic was automatically closed 90 days after the last reply. New replies are no longer allowed.