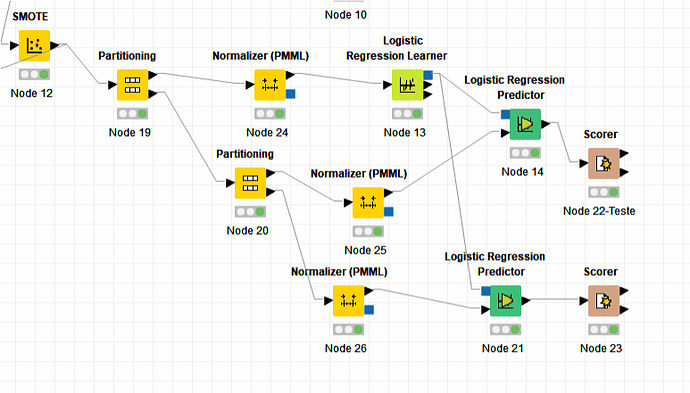
Guys
I’m a beginner and would like some help. I’m trying to reproduce the code below from python in Knime
but the results of train accuracy, dev accuracy, test accuracy and confusion matrix in logistic regression are not correct
Phyton country calling code
–Smote
col = ‘Category’
data = sample_data(df, col)
—Randomly distribute data into training, testing and validation classes. We use 60-20-20 distribution
un_training_x, training_y, un_testing_x, testing_y, un_validation_x, validation_y = split_random(data, percent_train=6
—Lets normalize our X data
training_x, testing_x, validation_x = normalize_data(un_training_x, un_testing_x, un_validation_x)
We can print the X data, to be sure that we have the normalized data in the range of -1 to 1
print(“X:”)
print_normalized_data(training_x, testing_x, validation_x)
print(“")
—Lets print the Y class, to be sure that we have a mix of positive and negative class
print(“Y”)
print_normalized_data(training_y, testing_y, validation_y)
print("”)
Logistic Regression
clf = sklearn.linear_model.LogisticRegressionCV(penalty=‘l2’, solver=‘lbfgs’,
print(clf.fit(training_x.T, training_y.T.reshape(training_x.shape[1],)))
LogisticRegressionCV(Cs=10, class_weight=None, cv=None, dual=False,
fit_intercept=True, intercept_scaling=1.0, max_iter=1000,
multi_class=‘ovr’, n_jobs=1, penalty=‘l2’, random_state=None,
refit=True, scoring=None, solver=‘lbfgs’, tol=0.0001,
verbose=1.0)
cc_dic = {}
cc_dic = analyze_results(training_x, training_y, validation_x, validation_y, testing_x, testing_y, “lr_sklearn”, None, clf, acc_dic)
Train accuracy: 96.61319073083779
Dev accuracy: 87.76595744680851
Test accuracy: 96.79144385026738
Confusion matrix of Testing Data:
[[86 5]
[ 1 95]]
Knime representation