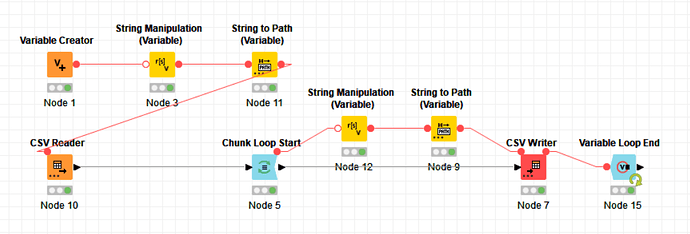
Hi guys, I made a simple solution with loops and files path to make it.
For the example, I used the Chunck Loop start, because it can split it in fixed numbers of rows or parts/groups, make it easier to manipulate as you wish.
Inside the loop session, I just build a string for the path with a counter, manipulating the information by the Interactive end loop session. and before the end loop node, I insert a write csv node to export the data for a file.
file_split.knwf (363.8 KB)
For the loop session, I’d like to break a part with 1.000 rows of data for a file.
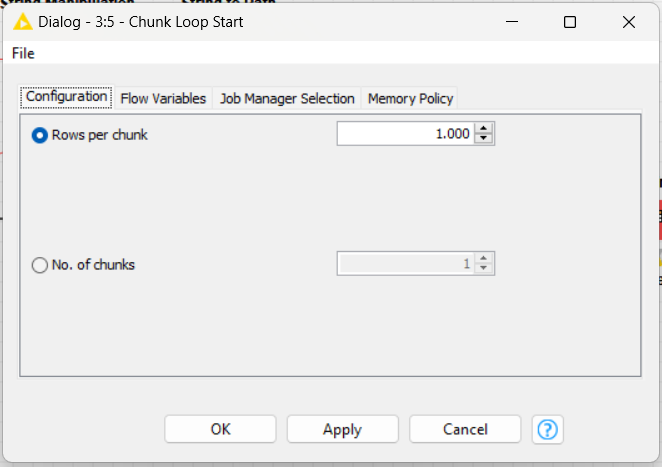
You can say that you’d like to break on 4 parts/chucks as you wish.
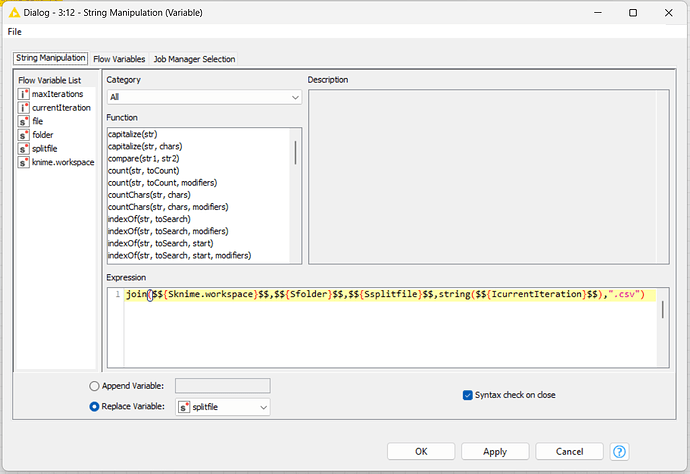
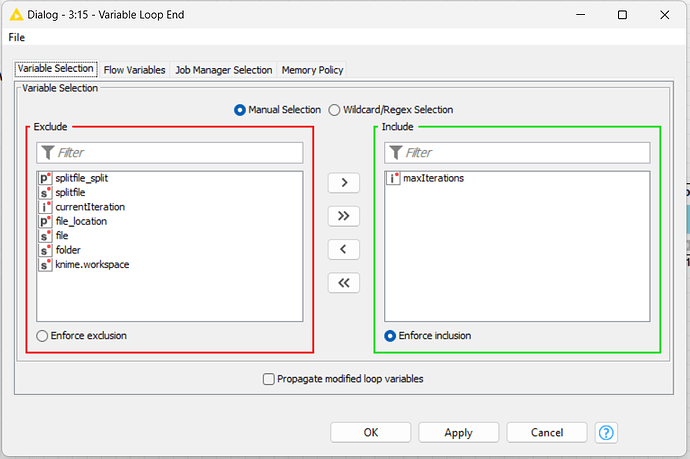
Inside the loop, I’ll use variables (“currentInteraction” and “maxInteraction”) to manipulate the file name result and control de end of the loop.
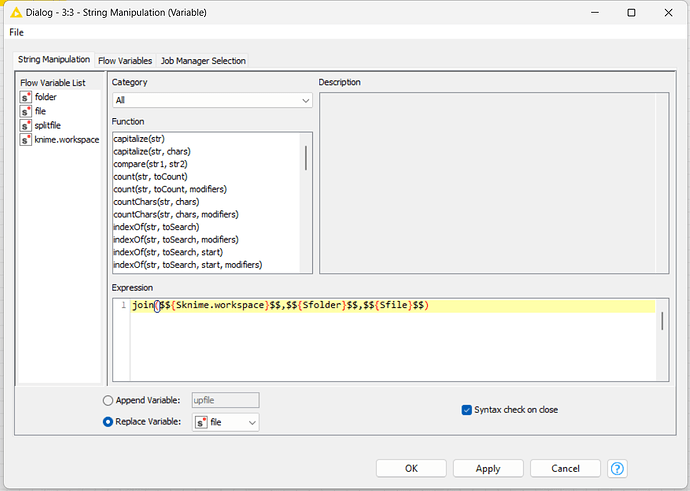
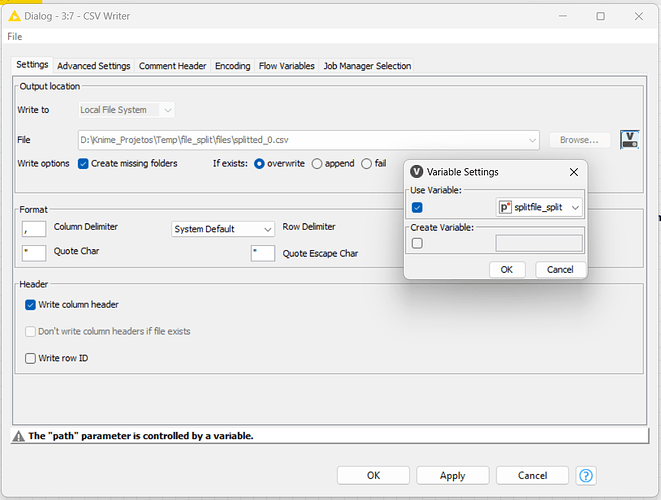
Expression: join($${Sknime.workspace}$$,$${Sfolder}$$,$${Ssplitfile}$$,string($${IcurrentIteration}$$),“.csv”)
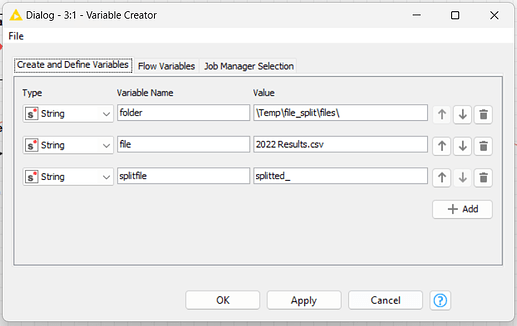
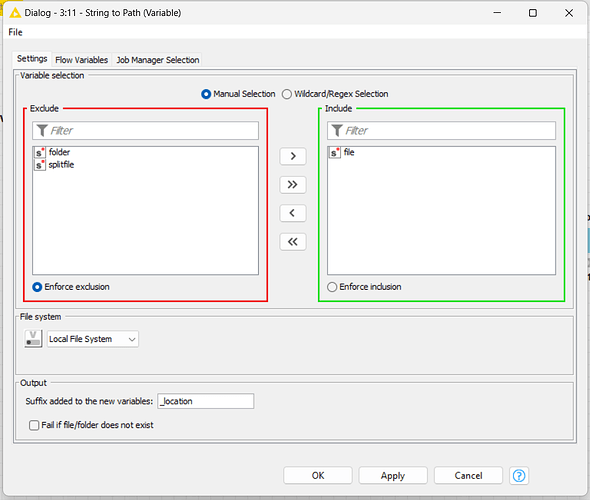
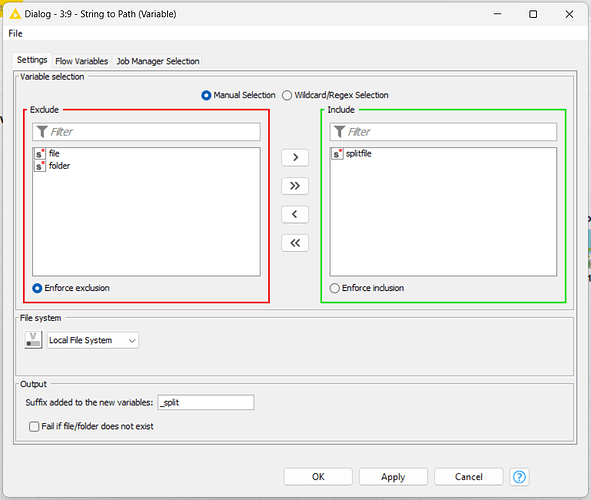
I need to save the new file to another variable, I called it as “splitfile”, then I create a new path for it too.
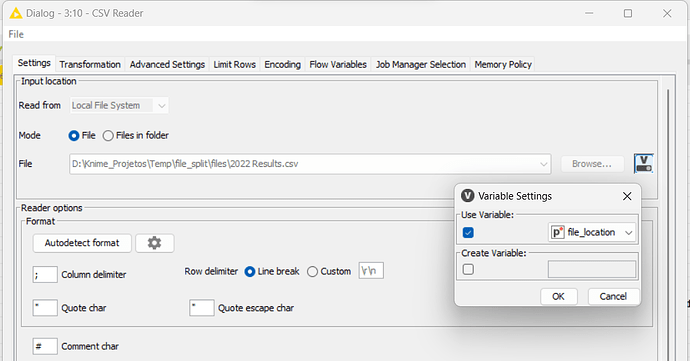
With the CSV Writer node, I’ll set the file path with the workflow variables, using the splitfile path indicator.
And for the ends, I used the Variables End loop node to check the MaxInteraction that used for this situation.
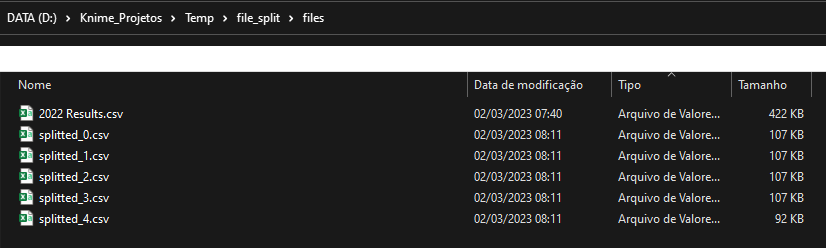
The result was this:
Can it solve your problem?
Tks,
Denis