Hello everyone,
I’m trying to extract all PAINS from a huge database (3.5M structures), and I have a text file with SMARTS strings for all PAINS reported in the literature. The issue is, no matter what, the progress on the execution of the SMARTS Query node gets stuck at 25% without any errors, and just doesn’t go forward.
I’m struggling to find a cause for this, and would be grateful if someone shared an insight on what might be happening.
Hi @kbisikalo
Welcome to the Knime forum.
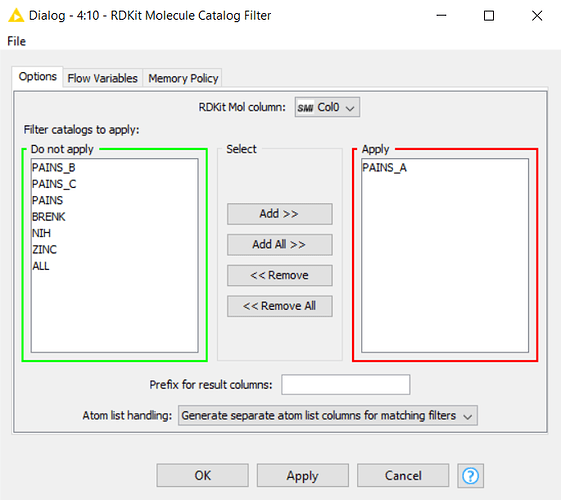
There is a node called “RDKit Molecule Catalog Filter” by RDKit which does the job of looking for PAINS (among other possible things). Would this help to achieve your task?
Are you doing the query using a database query node or importing first into KNIME the list of molecules and then doing the SMART query directly with KNIME ?
Would it be possible to share a minimalist workflow to better understand what you are doing so that we can help ?
Best regards,
Ael
Firstly, thank you for sharing “RDKit Molecule Catalog Filter” node with me, I will test it on a smaller set right now, and maybe it will eliminate my problems altogether!
I do have an sdf file of the entire database, and I have imported it directly using an sdf reader node. I have attached a workflow that I’m using, that have been tested succesfully on a smaller set (200k strucrures).Example_SMARTS_Query.knwf (10.1 KB)
Update: The “RDKit Molecule Catalog Filter” works for on my smaller test set, but is there a way to check what substructures are in what filter in this node? I may need a more presice control over the PAINS I extract, and would like to at least know what is being filtered.
I would recommend to use the “RDKit Molecule Substructure Filter” to do a search by SMARTS instead of the node you are using by CDK. You can use your SMART list of PAINS as input for the second input port and hence have full knowledge of what is filtered.
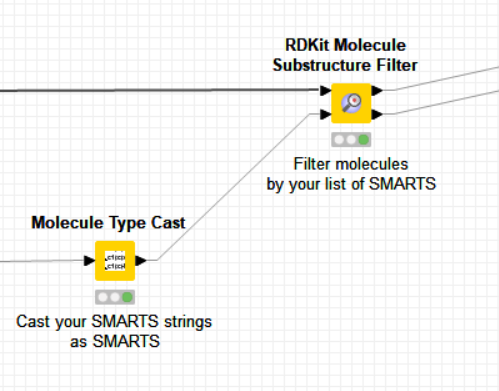
Hope this helps.
Ael
Thank you very much, I would launch the updated workflow for my db now!
Hi @kbisikalo
Many thanks for validating the answer.
As complementary information, this RDKit github link (rdkit/Code/GraphMol/FilterCatalog at master · rdkit/rdkit · GitHub) contains the files that most problably are used as PAINS references (pains_a.in, pains_b.in & pains_c.in) in the “RDKit Molecule Catalog Filter”, in case you would like to compare them with your own list of PAINS smarts.
Hope this helps.
Best wishes,
Ael
Hi,
This may also be useful: GitHub - PatWalters/rd_filters: A script to run structural alerts using the RDKit and ChEMBL.
I tend to use the RDKit Functional Group Filter node for this kind of stuff, because it allows you to read the Smarts patterns from a text file.
Cheers/Evert
Now that I have achieved what I was doing, I can add a few comments.
Probably the most helpful node to use in this case is the RDKit Catalogue filter, as it is the fastest of them all, and still gives pretty good info.
The RDKit Molecule Substructure filter turned out to be a weird one. On my smaller set, with my own SMARTS strings it worked just fine, but on the beg set it just refused to output data (0 matching molecules). The strangest part is that my test set is literally a diversity selection from the big one, with everything being the same.
And lastly, it turned out that the problem I was having with the SMARTS Query node (getting stuck at 25% forever) can be solved by throwing increasing amounts of computing power and time at it. Running it on a beefier pc overnight solved the issue and provided info that I needed.
This topic was automatically closed 7 days after the last reply. New replies are no longer allowed.