Hi,
I’m using the 3.7.0 Knime version under windows 10.
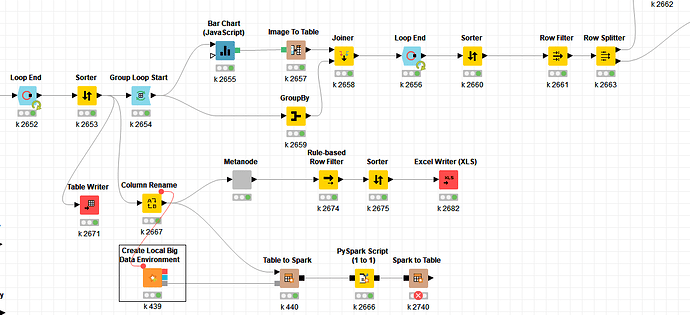
The node fail very quickly after the pyspark node.
I’m not using spark for production but only for development and test jobs. The table in entry count 3708 lines.
Could it come from the Pyspark node ? A dirty hidden instructions with a copy paste ?
The log file :
2019-01-07 00:37:42,072 : INFO : main : NodeLogger : : : #########################################################################################
2019-01-07 00:37:42,404 : INFO : main : NodeLogger : : : # #
2019-01-07 00:37:42,404 : INFO : main : NodeLogger : : : # Welcome to KNIME Analytics Platform v3.7.0.v201812041331 (Build December 06, 2018) #
2019-01-07 00:37:42,404 : INFO : main : NodeLogger : : : # Based on Eclipse, http://www.eclipse.org #
2019-01-07 00:37:42,404 : INFO : main : NodeLogger : : : # #
2019-01-07 00:37:42,404 : INFO : main : NodeLogger : : : #########################################################################################
2019-01-07 00:37:42,404 : INFO : main : NodeLogger : : : # #
2019-01-07 00:37:42,404 : INFO : main : NodeLogger : : : # Copyright by KNIME AG, Zurich, Switzerland and others. #
2019-01-07 00:37:42,404 : INFO : main : NodeLogger : : : # Website: http://www.knime.com #
2019-01-07 00:37:42,404 : INFO : main : NodeLogger : : : # E-mail: contact@knime.com #
2019-01-07 00:37:42,404 : INFO : main : NodeLogger : : : # #
2019-01-07 00:37:42,404 : INFO : main : NodeLogger : : : #########################################################################################
2019-01-07 00:37:42,404 : INFO : main : NodeLogger : : : # For more details see the KNIME log file: #
2019-01-07 00:37:42,404 : INFO : main : NodeLogger : : : # F:\knime_depot2.metadata\knime\knime.log
2019-01-07 00:37:42,404 : INFO : main : NodeLogger : : : #---------------------------------------------------------------------------------------#
2019-01-07 00:37:42,419 : INFO : main : NodeLogger : : : # logging date=Mon Jan 07 00:37:42 CET 2019
2019-01-07 00:37:42,419 : INFO : main : NodeLogger : : : # java.version=1.8.0_152
2019-01-07 00:37:42,419 : INFO : main : NodeLogger : : : # java.vm.version=25.152-b16
2019-01-07 00:37:42,419 : INFO : main : NodeLogger : : : # java.vendor=Oracle Corporation
2019-01-07 00:37:42,419 : INFO : main : NodeLogger : : : # os.name=Windows 10
2019-01-07 00:37:42,419 : INFO : main : NodeLogger : : : # os.arch=amd64
2019-01-07 00:37:42,419 : INFO : main : NodeLogger : : : # number of CPUs=8
2019-01-07 00:37:42,419 : INFO : main : NodeLogger : : : # assertions=off
2019-01-07 00:37:42,419 : INFO : main : NodeLogger : : : # host=vedmedik
2019-01-07 00:37:42,419 : INFO : main : NodeLogger : : : # username=FC2018
2019-01-07 00:37:42,419 : INFO : main : NodeLogger : : : # max mem=18098MB
2019-01-07 00:37:42,419 : INFO : main : NodeLogger : : : # application=org.knime.product.KNIME_APPLICATION
2019-01-07 00:37:42,419 : INFO : main : NodeLogger : : : # ID=01-196c30c2d7ff34a1
2019-01-07 00:37:42,419 : INFO : main : NodeLogger : : : #########################################################################################
2019-01-07 00:37:46,827 : DEBUG : main : DatabaseConnectionSettings : : : Settings database timeout to 15 seconds
2019-01-07 00:37:47,030 : DEBUG : KNIME-Worker-1 : KnimeEnterpriseFileSystemPlugin : : : Started KNIME Enterprise Remote File System plug-in
2019-01-07 00:37:47,046 : INFO : KNIME-Worker-1 : ExplorerMountTable : : : Mounted Explorer Temp Space ‘knime-temp-space’ - com.knime.explorer.tempspace
2019-01-07 00:37:48,209 : DEBUG : main : DatabaseConnectionSettings : : : Database concurrency (sync via database connection) is true.
2019-01-07 00:37:48,209 : DEBUG : main : KNIMECorePlugin : : : Setting KNIME max thread count to 16
2019-01-07 00:37:48,209 : DEBUG : main : KNIMECorePlugin : : : Setting KNIME temp dir to “D:\knime_temp”
2019-01-07 00:48:43,098 : ERROR : KNIME-Worker-8 : Node : Spark to Table : 0:2740 : Execute failed: Accept timed out (SocketTimeoutException)
I join the Pyspark instructions in any case :
System imports
import sys
from pyspark.mllib.common import _py2java, _java2py
from pyspark import SparkContext, SparkConf, SparkFiles
from pyspark.sql import SQLContext
from pyspark.serializers import PickleSerializer
from pyspark.sql import SparkSession
from pyspark.sql.types import *
from pyspark.java_gateway import launch_gateway
from pyspark.profiler import BasicProfiler
from collections import OrderedDict
class Exchanger(object):
_spark = None
_jexchange = None
def init (self):
_gateway = launch_gateway()
_jvm = _gateway.jvm
self._jexchange = _jvm.org.knime.bigdata.spark2_4.jobs.scripting.python.PySparkDataExchanger.SINGLETON_INSTANCE
jcontext = self._jexchange.getContext()
jsession = self._jexchange.getSession()
jconf = jcontext.getConf()
conf = SparkConf(loadDefaults=False, _jvm=_jvm, _jconf=jconf)
sparkCon = SparkContext(None, None, None, None, None, 0, PickleSerializer(), conf, _gateway, jcontext, BasicProfiler)
self._spark = SparkSession(sparkCon,jsession)
def getDataFrame(self, name):
javain = self._jexchange.getDataFrame(name)
df_in = _java2py(self._spark, javain)
return df_in
def addDataFrame(self, name, df):
jdf = _py2java(self._spark, df)
self._jexchange.addDataFrame(name,jdf)
def addObject(self, name, obj):
jobj = _py2java(self._spark, obj)
self._jexchange.add(name,jobj)
def getObject(self, name):
javain = self._jexchange.get(name)
obj = _java2py(self._spark, javain)
return obj
def getSparkSession(self):
return self._spark
Your custom imports:
#import statistics
from pyspark.mllib.stat import Statistics
import pyspark.sql.functions as F
from pyspark.sql.types import *
from pyspark.sql.window import Window
from pyspark.sql.functions import date_format
from scipy.stats import chi2
Flowvariables
Your custom global variables:
Initialization of Spark environment
if name == “main”:
_exchanger = Exchanger()
spark = _exchanger.getSparkSession()
# Get data from jvm
dataFrame1 = _exchanger.getDataFrame(“xxx_dataFrame1”)
#End of initialization
# Custom pySpark code
# SparkSession can be used with variable spark
# The input dataFrame(s): [dataFrame1]
# The output dataFrame(s) must be: [resultDataFrame1]
#resultDataFrame1 = dataFrame1
#@udf('double')
def chi2proba(x):
"""calculate chi2 probability from a value
"""
try: p = float(chi2.cdf(x, df=1, loc=0, scale=1))
except: p = 1
return p
chi2proba = F.udf(lambda z: chi2proba(z), DoubleType())
def chi2proba_table(df, distance):
"""approximate the chi2 probability give a result similar to chi2proba without using udf
only a join on a reference table
"""
df = df.withColumn("dist_", F.round(distance, 2))
df_proba = spark.sql("select dist_,proba_ from fab_geokpi.dist_to_proba")
df = df.join(df_proba, ["dist_"], how='left')
df = df.withColumn("proba1", F.when("dist_" > F.lit(20), 1)\
.otherwise(df["proba_"])).drop("proba_")
df = df.withColumnRenamed("proba1", "proba").drop("dist_")
return df
##########################################
### utilitaires divers
#test UDF brouillons etc...
##########################################
def concatenate_dynamic(df, groups):
"""concatenate columns generate a unique key for grouping
for string column only
internal usage - do not deliver final result
"""
for idcol, colname in enumerate(groups):
if idcol == 0:
origine = df.withColumn("key_seg", df[colname])
else:
origine = origine.withColumn("key_seg", F.concat(origine["key_seg"], origine[colname]))
return origine
def add_day(df, day_column="day"):
"""help to insert day description value into a table
to know what field you can had look at
http://www.dataclear.club/index.php?post/2017/05/28/Une-table-de-gestion-des-dates
"""
df = df.withColumn("date_string", date_format(F.col(day_column), "YYYY-MM-dd"))
req = "select date_string, jours_semaine,mois_string,ferie from fab_geokpi.s_calendrier"
df_cal = spark.sql(req)
df = df.join(df_cal, df["date_string"] == df_cal["date_string"])
df = df.drop("date_string")
return df
################################################
### Fonctions applicables sur une colonne entiere
################################################
def flag_metier(df, target_column, threshold=1, annule='n'):
"""mandatory classification rules overcoming automatized chi2 rules
"""
if annule == 'n':
df = df.withColumn('mandatory', \
F.when(df[target_column] == 0, 1)
.when(F.col(target_column).isNull(), 2)
.when(df[target_column] < 0, 3)
.when(((df[target_column] > 0) & (df[target_column] < F.lit(threshold))), 4)
.otherwise(0))
else: df = df.withColumn('mandatory', F.lit(0))
return df
def get_median(df, target_column):
"""return limit of first and third quartiles of a distribution
for big big data increase the third parameter (default here 0)
ina [0,1] span to gain calculation time
internal usage - do not deliver final result
"""
df_res = df.approxQuantile(target_column, [0.5], 0)
return df_res
def get_median_by_key(df, target_column, group_columns):
"""return limit of first and third quartiles of a distribution
internal usage - do not deliver final result
"""
grp_window = Window.partitionBy(group_columns)
median = F.expr('percentile_approx('+target_column+', 0.50)')
df_res = df.withColumn("median", median.over(grp_window))
return df_res
########## bloc mahalanobis ###########################
def mahalanobis_one(df, target_column, option="calc", limit_max=30):
"""caclulate the mahalanobis distance between a set of values and its median
when there is no group_columns vector defined
"""
grp_window = Window.partitionBy()
df = flag_metier(df=df, target_column=target_column)
median = get_median(df=df, target_column=target_column)[0]
df = df.withColumn("median", F.lit(median))
df = df.withColumn("eff", F.count(target_column).over(grp_window))
df = df.withColumn("deviation1", F.pow(df[target_column]-df["median"], 2))
df = df.withColumn("deviation2", F.sum(df["deviation1"]).over(grp_window))
df = df.withColumn("deviation3", df["deviation2"]/(df["eff"]-F.lit(1)))
df = df.withColumn("deviation", F.sqrt(df["deviation3"])).fillna(0)
df = df.withColumn("maxi", F.max(df[target_column]).over(grp_window))
df = df.withColumn("criteria",\
F.sqrt(F.pow((df[target_column]-df["median"]), 2)/F.pow(df["deviation"], 2)))
if option == "calc":
df = df.withColumn("proba", F.when((df["maxi"] > F.lit(limit_max)), chi2proba(df.criteria))\
.otherwise(F.lit(0))).fillna(0)
df = df.drop("maxi")
elif option == "table":
df = chi2proba_table(df=df, distance="criteria")
df = df.withColumn("proba1", F.coalesce(df.proba, F.lit(0)))
df = df.withColumn("proba", F.when(df["maxi"] > F.lit(limit_max), df["proba1"])\
.otherwise(F.lit(0))).drop("proba1").drop("maxi")
df = df.select([c for c in df.columns if c not in {"deviation1", "deviation2", "deviation3", "deviation", "criteria", "eff"}])
return df
def mahalanobis(df, target_column, group_columns, option="calc", limit_max=30):
"""calculate the mahalanobis distance between a set of values and its median
and add the corresponding probability according to the limit_max criteria
"""
if group_columns == []:
df = mahalanobis_one(df=df, target_column=target_column, limit_max=limit_max)
else:
df = concatenate_dynamic(df=df, groups=group_columns)
grp_window = Window.partitionBy("key_seg")
df = df.withColumn("eff", F.count(target_column).over(grp_window))
df = get_median_by_key(df=df, target_column=target_column, group_columns=["key_seg"])
df = df.withColumn("deviation1", F.pow(df[target_column]-df["median"], 2))
df = df.withColumn("deviation2", F.sum(df["deviation1"]).over(grp_window))
df = df.withColumn("deviation3", df["deviation2"]/(df["eff"]-F.lit(1)))
df = df.withColumn("deviation", F.sqrt(df["deviation3"])).fillna(0)
df = df.withColumn("criteria", F.sqrt(F.pow((df[target_column]-df["median"]), 2)/F.pow(df["deviation"], 2)))
df = df.withColumn("maxi", F.max(df[target_column]).over(grp_window))
if option == "calc":
df = df.withColumn("proba", F.when(df["maxi"] > F.lit(limit_max), chi2proba(df.criteria))\
.otherwise(F.lit(0))).fillna(0)
df = df.drop("maxi")
elif option == "table":
df = chi2proba_table(df=df, distance="criteria")
df = df.withColumn("proba1", F.coalesce(df.proba, F.lit(0)))
df = df.withColumn("proba", F.when(df["maxi"] > F.lit(limit_max), df["proba1"])\
.otherwise(F.lit(0))).drop("proba1").drop("maxi")
df = df.select([c for c in df.columns if c not in {"deviation1", "deviation2", "deviation3", "deviation", "criteria", "eff", "key_seg"}])
return df
############ Fonction generale ##########
def anomalie_helper(df, target_column, group_columns, threshold1, threshold2=0, option="calc", limit_max=30):
"""calculate the scores and alerting flags for one columns
"""
df = flag_metier(df=df, target_column=target_column, threshold=threshold1)
df = mahalanobis(df=df, target_column=target_column, group_columns=group_columns, option=option, limit_max=limit_max)
df = df.withColumn("probability", F.when((df["mandatory"] > 0), 1).otherwise(df["proba"])).drop("proba")
if threshold2 > 0 and threshold2 <= 1:
df = df.withColumn("verdict",
F.when((df["probability"] > F.lit(threshold2)) & (df[target_column] < df["median"]), "-")
.when((df["probability"] > F.lit(threshold2)) & (df[target_column] >= df["median"]), "+")
.when(df['mandatory'] > F.lit(0),"XXXX")
.otherwise("ok"))
elif threshold2 == 0:
df = df.withColumn("verdict",
F.when(df['mandatory'] > F.lit(0), "XXXX")
.when((df["probability"] > F.lit(0.7)) & (df["probability"] <= F.lit(0.8)) & (df[target_column] < df["median"]), "-")
.when((df["probability"] > F.lit(0.8)) & (df["probability"] <= F.lit(0.9)) & (df[target_column] < df["median"]), "- -")
.when((df["probability"] > F.lit(0.9)) & (df["probability"] <= F.lit(0.95)) & (df[target_column] < df["median"]), "- - -")
.when((df["probability"] > F.lit(0.95)) & (df["probability"] <= F.lit(1)) & (df[target_column] < df["median"]), "- - - -")
.when((df["probability"] > F.lit(0.7)) & (df["probability"] <= F.lit(0.8)) & (df[target_column] >= df["median"]), "+")
.when((df["probability"] > F.lit(0.8)) & (df["probability"] <= F.lit(0.9)) & (df[target_column] >= df["median"]), "+ +")
.when((df["probability"] > F.lit(0.9)) & (df["probability"] <= F.lit(0.95)) & (df[target_column] >= df["median"]), "+ + +")
.when((df["probability"] > F.lit(0.95)) & (df["probability"] <= F.lit(1)) & (df[target_column] >= df["median"]), "+ + + +")
.otherwise("ok"))
df = df.drop("median").drop("mandatory")
return df
def anomalie_helper_multicolumn(df, target_columns, group_columns, thresholds1=[10, 3000], thresholds2=[0, 0], option="calc", limit_max=[30, 30]):
"""calculate the scores and alerting flags for many columns
"""
for i, x_col in enumerate(target_columns):
df = anomalie_helper(df=df, target_column=x_col, group_columns=group_columns,threshold1=thresholds1[i], threshold2=thresholds2[i], option=option, limit_max=limit_max[i])
df = df.withColumnRenamed("verdict", "verdict_"+x_col).withColumnRenamed("probability", "probability_"+x_col)
return df
df = dataFrame1
#df_test = add_day(df=df)
resultDataFrame1 = anomalie_helper_multicolumn(df=df, group_columns=["jour_semaine","hour"],target_columns=["nbposition"], thresholds1=[0,0,0], thresholds2=[0,0,0], limit_max=[1,1,1], option="calc")
# End of user code
# Send data to jvm
Happy new year to you !