Can survival analysis / Cox proportional hazards regression be performed using Knime?
Hi,
this is possible via Python scripting nodes : https://github.com/CamDavidsonPilon/lifelines
Or R nodes https://cran.r-project.org/web/packages/survival/index.html
Best regards
Fabien
Is possible to perform logrank test in survival analysis (Kaplan Meier node) in Knime without R or Python?
Hello,
I cannot find the survival analysis node in my Knime program . ¿Any could help me? Thanks
The user referred to python and R so maybe you like to go with these nodes
Hello KNIME community,
We’re conducting research where we would like to search scientific studies for any study containing Kaplan-Meier survival curves for a specific disease.
The problem is that this data is not included in the titles or abstracts of studies, but is in the full text. Is there a method where we could search PubMed or Scopus for full text containing Kaplan-Meier curves without having to screen thousands of studies that have the word survival in the title and/or abstract?
(i.e. a tool that could help us search for the word Kaplan-Meier or overall survival inside the full texts?)
Thanks in advance,
Baher Elezbawy
Maybe there is an API which allows searching for key terms even if they are not in the title or abstract?
Edit: sth like this?
hi @baherelezbawy
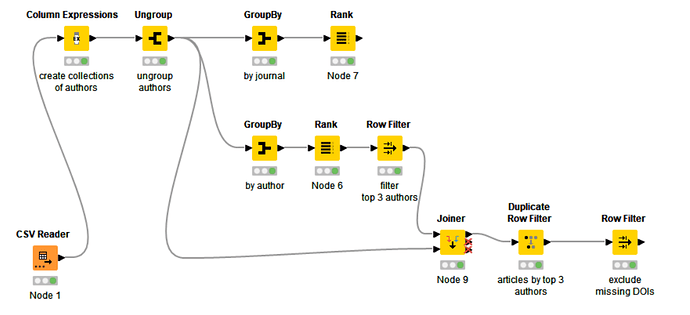
I don’t think it is possible to search the full texts via API or web interface, at least on PubMed, which is, as long as I know, a bibliographic database, not a fulltext one. Perhaps it would be helpful to do some very targeted research on PubMed, download the results as a csv file, then use KNIME for example to select the main authors and journals and focus on them. I’ve tried it myself, searching “Kaplan-Meier” in all fields, then refining the search using some filters, finally downloading the csv file (7313 rows). This is an example workflow where I rank authors and journals according to the number of articles.
KNIME_project.knwf (146.5 KB)
Using the DOIs you can reach the fulltext. For example, the paper with DOI 10.2217/fon-2021-0101 can be found here: https://dx.doi.org/10.2217/fon-2021-0101
Thank you so much Duristef
Thank you so much Daniel
On second thoughts, this is not a good idea, mainly because of the author’s name abbreviation (e.g. “Zhang Y”).
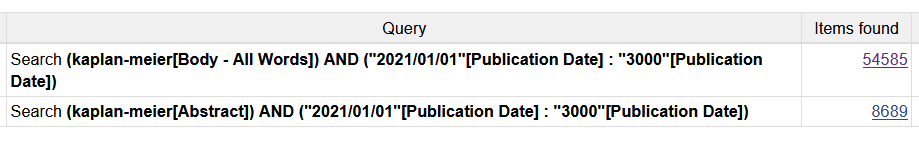
PubMed is a bibliographic database, but PubMed Central is fulltext and allows fulltext searching
Results (including full author names) can be downloaded in text or XML format (which is really heavy).
I’ve downloaded a small dataset (291 articles) in XML format and this is the new workflow
PubMedCentral_search_xml.knwf (35.0 KB)
The last workflow is based on the Vernalis node “European PubMed Central Advanced Search” (close but no cigar: fulltext not available AFAIK)
EuroPubMed_search_xml.knwf (36.9 KB)
Hope these workflow can help you, at least as suggestions. I think I have learned more myself than I have been able to help