Hi,
I have some question about the workflow in handout about openms.
a)Does the workflow of Label-free quantification and identification workflow for metabolites(Figure 36)the relation with the Metabolite Adduct Decharger adduct grouping workflow.
b) When I process the data I wonder I should choose which workflow.
c) The result of MS2 information and MS1 should I combine the result as my final data ,then I use the final data to visualize.
Hi,
a) MetaboliteAdductDecharging is just an optional, additional step, that you can do to “cluster” features from the same metabolite with different adducts. It usually improves results but takes additional runtime.
b) Depends on which data you have.
c) Yes if you have data-dependent acquired MS2 fragment scans it would be helpful to use e.g. Sirius+CSIFingerID or spectral matching to narrow down candidates per MS1 feature. MS2 scans from untargeted data-independent acquisition currently cannot be used by the OpenMS plugin. They have to be ignored. For targeted DIA, you can use the DIAMetAlyzer workflow or the OpenSwath for metabolomics tutorial.
Thank you very much.
Hi,
Yeah, however, there is an new error.
Execute failed: Failed to execute node MetaboliteAdductDecharger
ERROR MetaboliteAdductDecharger 3:266 Failing process stdout: [Adding neutral: ---------- Adduct -----------------, Charge: 0, Amount: 1, MassSingle: -18.0106, Formula: H-2O-1, log P: -2.99573, ]
ERROR MetaboliteAdductDecharger 3:266 Compomer: Da -14.9434; q_net 2; logP -8.9872[[ (K1) → (H-2O-1H1H4N1Na1) ]], Compomer: Da -13.9594; q_net 2; logP -5.52146[[ (K1) → (H2Na1) ]], Compomer: Da -11.0406; q_net 2; logP -7.6009[[ (H4N1) → (H-2O-1H2Na1) ]], Compomer: Da -9.98801; q_net 2; logP -8.9872[[ (K1) → (H-2O-1H1Na2) ]], Compomer: Da -3.92486; q_net 2; logP -8.07091[[ (Na1) → (H-2O-1H1H8N2) ]], Compomer: Da -2.94084; q_net 2; logP -4.60517[[ (Na1) → (H2H4N1) ]], Compomer: Da -2.87225; q_net 2; logP -9.4572[[ (K1) → (H-2O-1H12N3) ]], Compomer: Da -1.95683; q_net 2; logP -7.1309[[ (H-2O-1Na1) → (H3) ]], Compomer: Da -1.88823; q_net 2; logP -5.99146[[ (K1) → (H1H8N2) ]], Compomer: Da -0.904215; q_net 2; logP -8.51719[[ (H-2O-1K1) → (H2H4N1) ]], Compomer: Da -0.0220747; q_net 2; logP -8.51719[[ (Na1) → (H-2O-1H2K1) ]], Compomer: Da 1.03054; q_net 2; logP -5.29832[[ () → (H-2O-1H1H4N1) ]], Compomer: Da 2.01455; q_net 2; logP -1.83258[[ () → (H2) ]], Compomer: Da 2.08315; q_net 2; logP -9.4572[[ (K1) → (H-2O-1H8N2Na1) ]], Compomer: Da 2.99857; q_net 2; logP -7.1309[[ (H-2O-1H4N1) → (H3) ]], Compomer: Da 3.06716; q_net 2; logP -5.99146[[ (K1) → (H1H4N1Na1) ]], Compomer: Da 4.05118; q_net 2; logP -8.51719[[ (H-2O-1K1) → (H2Na1) ]], Compomer: Da 4.93332; q_net 2; logP -8.51719[[ (H4N1) → (H-2O-1H2K1) ]], Compomer: Da 5.98593; q_net 2; logP -5.29832[[ () → (H-2O-1H1Na1) ]], Compomer: Da 6.96995; q_net 2; logP -4.60517[[ (H4N1) → (H2Na1) ]], Compomer: Da 7.03854; q_net 2; logP -9.4572[[ (K1) → (H-2O-1H4N1Na2) ]], Compomer: Da 8.02256; q_net 2; logP -5.99146[[ (K1) → (H1Na2) ]], Compomer: Da 10.9413; q_net 2; logP -8.07091[[ (H4N1) → (H-2O-1H1Na2) ]], Compomer: Da 11.9939; q_net 2; logP -9.4572[[ (K1) → (H-2O-1Na3) ]], Compomer: Da 13.1017; q_net 2; logP -8.54091[[ (Na1) → (H-2O-1H12N3) ]], Compomer: Da 14.0857; q_net 2; logP -5.07517[[ (Na1) → (H1H8N2) ]], Compomer: Da 15.0697; q_net 2; logP -7.6009[[ (H-2O-1Na1) → (H2H4N1) ]], Compomer: Da 15.1383; q_net 2; logP -6.46147[[ (K1) → (H12N3) ]], Compomer: Da 16.1223; q_net 2; logP -8.9872[[ (H-2O-1K1) → (H1H8N2) ]], Compomer: Da 17.0045; q_net 2; logP -8.9872[[ (Na1) → (H-2O-1H1H4N1K1) ]], Compomer: Da 17.9885; q_net 2; logP -5.52146[[ (Na1) → (H2K1) ]], Compomer: Da 18.0571; q_net 2; logP -5.76832[[ () → (H-2O-1H8N2) ]], Compomer: Da 19.0411; q_net 2; logP -2.30259[[ () → (H1H4N1) ]], Compomer: Da 20.0251; q_net 2; logP -4.82831[[ (H-2O-1) → (H2) ]], Compomer: Da 20.0937; q_net 2; logP -6.46147[[ (K1) → (H8N2Na1) ]], Compomer: Da 21.0777; q_net 2; logP -8.9872[[ (H-2O-1K1) → (H1H4N1Na1) ]], Compomer: Da 21.9599; q_net 2; logP -6.21461[[ () → (H-2O-1H1K1) ]], Compomer: Da 22.9439; q_net 2; logP -5.52146[[ (H4N1) → (H2K1) ]], Compomer: Da 23.0125; q_net 2; logP -5.76832[[ () → (H-2O-1H4N1Na1) ]], Compomer: Da 23.9965; q_net 2; logP -2.30259[[ () → (H1Na1) ]], Compomer: Da 24.9805; q_net 2; logP -7.6009[[ (H-2O-1H4N1) → (H2Na1) ]], Compomer: Da 25.0491; q_net 2; logP -6.46147[[ (K1) → (H4N1Na2) ]], Compomer: Da 26.0331; q_net 2; logP -8.9872[[ (H-2O-1K1) → (H1Na2) ]], Compomer: Da 26.9153; q_net 2; logP -8.9872[[ (H4N1) → (H-2O-1H1K1Na1) ]], Compomer: Da 27.9679; q_net 2; logP -5.76832[[ () → (H-2O-1Na2) ]], Compomer: Da 28.9519; q_net 2; logP -5.07517[[ (H4N1) → (H1Na2) ]], Compomer: Da 30.0045; q_net 2; logP -6.46147[[ (K1) → (Na3) ]], Compomer: Da 31.1123; q_net 2; logP -5.54518[[ (Na1) → (H12N3) ]], Compomer: Da 32.0963; q_net 2; logP -8.07091[[ (H-2O-1Na1) → (H1H8N2) ]], Compomer: Da 32.9233; q_net 2; logP -8.54091[[ (H4N1) → (H-2O-1Na3) ]], Compomer: Da 33.1489; q_net 2; logP -9.4572[[ (H-2O-1K1) → (H12N3) ]], Compomer: Da 34.031; q_net 2; logP -9.4572[[ (Na1) → (H-2O-1H8N2K1) ]], Compomer: Da 35.015; q_net 2; logP -5.99146[[ (Na1) → (H1H4N1K1) ]], Compomer: Da 35.0836; q_net 2; logP -8.07091[[ (H1) → (H-2O-1H12N3) ]], Compomer: Da 35.9991; q_net 2; logP -8.51719[[ (H-2O-1Na1) → (H2K1) ]], Compomer: Da 36.0677; q_net 2; logP -2.77259[[ () → (H8N2) ]], Compomer: Da 37.0517; q_net 2; logP -5.29832[[ (H-2O-1) → (H1H4N1) ]], Compomer: Da 37.9338; q_net 2; logP -9.90349[[ (Na1) → (H-2O-1H1K2) ]], Compomer: Da 38.1043; q_net 2; logP -9.4572[[ (H-2O-1K1) → (H8N2Na1) ]], Compomer: Da 38.9864; q_net 2; logP -6.68461[[ () → (H-2O-1H4N1K1) ]], Compomer: Da 39.9704; q_net 2; logP -3.21888[[ () → (H1K1) ]], Compomer: Da 40.039; q_net 2; logP -8.07091[[ (H1) → (H-2O-1H8N2Na1) ]], Compomer: Da 40.9545; q_net 2; logP -8.51719[[ (H-2O-1H4N1) → (H2K1) ]], Compomer: Da 41.023; q_net 2; logP -2.77259[[ () → (H4N1Na1) ]], Compomer: Da 42.0071; q_net 2; logP -5.29832[[ (H-2O-1) → (H1Na1) ]], Compomer: Da 42.8892; q_net 2; logP -9.90349[[ (H4N1) → (H-2O-1H1K2) ]], Compomer: Da 43.0597; q_net 2; logP -9.4572[[ (H-2O-1K1) → (H4N1Na2) ]], Compomer: Da 43.9418; q_net 2; logP -6.68461[[ () → (H-2O-1K1Na1) ]], Compomer: Da 44.9258; q_net 2; logP -5.99146[[ (H4N1) → (H1K1Na1) ]], Compomer: Da 44.9944; q_net 2; logP -8.07091[[ (H1) → (H-2O-1H4N1Na2) ]], Compomer: Da 45.9784; q_net 2; logP -2.77259[[ () → (Na2) ]], Compomer: Da 46.9625; q_net 2; logP -8.07091[[ (H-2O-1H4N1) → (H1Na2) ]], Compomer: Da 48.0151; q_net 2; logP -9.4572[[ (H-2O-1K1) → (Na3) ]], Compomer: Da 48.8972; q_net 2; logP -9.4572[[ (H4N1) → (H-2O-1K1Na2) ]], Compomer: Da 49.1228; q_net 2; logP -8.54091[[ (H-2O-1Na1) → (H12N3) ]], Compomer: Da 49.9498; q_net 2; logP -8.07091[[ (H1) → (H-2O-1Na3) ]], Compomer: Da 50.9338; q_net 2; logP -5.54518[[ (H4N1) → (Na3) ]], Compomer: Da 52.0416; q_net 2; logP -6.46147[[ (Na1) → (H8N2K1) ]], Compomer: Da 53.0256; q_net 2; logP -8.9872[[ (H-2O-1Na1) → (H1H4N1K1) ]], Compomer: Da 53.0942; q_net 2; logP -5.07517[[ (H1) → (H12N3) ]], Compomer: Da 54.0782; q_net 2; logP -5.76832[[ (H-2O-1) → (H8N2) ]], Compomer: Da 54.9604; q_net 2; logP -10.3735[[ (Na1) → (H-2O-1H4N1K2) ]], Compomer: Da 55.9444; q_net 2; logP -6.90776[[ (Na1) → (H1K2) ]], Compomer: Da 56.013; q_net 2; logP -8.9872[[ (H1) → (H-2O-1H8N2K1) ]], Compomer: Da 56.997; q_net 2; logP -3.68888[[ () → (H4N1K1) ]], Compomer: Da 57.981; q_net 2; logP -6.21461[[ (H-2O-1) → (H1K1) ]], Compomer: Da 58.0496; q_net 2; logP -5.07517[[ (H1) → (H8N2Na1) ]], Compomer: Da 59.0336; q_net 2; logP -5.76832[[ (H-2O-1) → (H4N1Na1) ]], Compomer: Da 59.9158; q_net 2; logP -7.6009[[ () → (H-2O-1K2) ]], Compomer: Da 60.8998; q_net 2; logP -6.90776[[ (H4N1) → (H1K2) ]], Compomer: Da 60.9684; q_net 2; logP -8.9872[[ (H1) → (H-2O-1H4N1K1Na1) ]], Compomer: Da 61.9524; q_net 2; logP -3.68888[[ () → (K1Na1) ]], Compomer: Da 62.9364; q_net 2; logP -8.9872[[ (H-2O-1H4N1) → (H1K1Na1) ]], Compomer: Da 63.005; q_net 2; logP -5.07517[[ (H1) → (H4N1Na2) ]], Compomer: Da 63.989; q_net 2; logP -5.76832[[ (H-2O-1) → (Na2) ]], Compomer: Da 64.8711; q_net 2; logP -10.3735[[ (H4N1) → (H-2O-1K2Na1) ]], Compomer: Da 65.9238; q_net 2; logP -8.9872[[ (H1) → (H-2O-1K1Na2) ]], Compomer: Da 66.9078; q_net 2; logP -6.46147[[ (H4N1) → (K1Na2) ]], Compomer: Da 67.9604; q_net 2; logP -5.07517[[ (H1) → (Na3) ]], Compomer: Da 68.9444; q_net 2; logP -8.54091[[ (H-2O-1H4N1) → (Na3) ]], Compomer: Da 70.0522; q_net 2; logP -9.4572[[ (H-2O-1Na1) → (H8N2K1) ]], Compomer: Da 71.1048; q_net 2; logP -8.07091[[ (H-2O-1H1) → (H12N3) ]], Compomer: Da 72.9709; q_net 2; logP -7.37776[[ (Na1) → (H4N1K2) ]], Compomer: Da 73.9549; q_net 2; logP -9.90349[[ (H-2O-1Na1) → (H1K2) ]], Compomer: Da 74.0235; q_net 2; logP -5.99146[[ (H1) → (H8N2K1) ]], Compomer: Da 75.0075; q_net 2; logP -6.68461[[ (H-2O-1) → (H4N1K1) ]], Compomer: Da 75.8897; q_net 2; logP -11.2898[[ (Na1) → (H-2O-1K3) ]], Compomer: Da 76.0602; q_net 2; logP -8.07091[[ (H-2O-1H1) → (H8N2Na1) ]], Compomer: Da 76.9423; q_net 2; logP -9.90349[[ (H1) → (H-2O-1H4N1K2) ]], Compomer: Da 77.9263; q_net 2; logP -4.60517[[ () → (K2) ]], Compomer: Da 78.9103; q_net 2; logP -9.90349[[ (H-2O-1H4N1) → (H1K2) ]], Compomer: Da 78.9789; q_net 2; logP -5.99146[[ (H1) → (H4N1K1Na1) ]], Compomer: Da 79.9629; q_net 2; logP -6.68461[[ (H-2O-1) → (K1Na1) ]], Compomer: Da 80.8451; q_net 2; logP -11.2898[[ (H4N1) → (H-2O-1K3) ]], Compomer: Da 81.0156; q_net 2; logP -8.07091[[ (H-2O-1H1) → (H4N1Na2) ]], Compomer: Da 81.8977; q_net 2; logP -9.90349[[ (H1) → (H-2O-1K2Na1) ]], Compomer: Da 82.8817; q_net 2; logP -7.37776[[ (H4N1) → (K2Na1) ]], Compomer: Da 83.9343; q_net 2; logP -5.99146[[ (H1) → (K1Na2) ]], Compomer: Da 84.9183; q_net 2; logP -9.4572[[ (H-2O-1H4N1) → (K1Na2) ]], Compomer: Da 85.971; q_net 2; logP -8.07091[[ (H-2O-1H1) → (Na3) ]], Compomer: Da 90.9815; q_net 2; logP -10.3735[[ (H-2O-1Na1) → (H4N1K2) ]], Compomer: Da 92.0341; q_net 2; logP -8.9872[[ (H-2O-1H1) → (H8N2K1) ]], Compomer: Da 93.9003; q_net 2; logP -8.29405[[ (Na1) → (K3) ]], Compomer: Da 94.9529; q_net 2; logP -6.90776[[ (H1) → (H4N1K2) ]], Compomer: Da 95.9369; q_net 2; logP -7.6009[[ (H-2O-1) → (K2) ]], Compomer: Da 96.9895; q_net 2; logP -8.9872[[ (H-2O-1H1) → (H4N1K1Na1) ]], Compomer: Da 97.8716; q_net 2; logP -10.8198[[ (H1) → (H-2O-1K3) ]], Compomer: Da 98.8556; q_net 2; logP -8.29405[[ (H4N1) → (K3) ]], Compomer: Da 99.9083; q_net 2; logP -6.90776[[ (H1) → (K2Na1) ]], Compomer: Da 100.892; q_net 2; logP -10.3735[[ (H-2O-1H4N1) → (K2Na1) ]], Compomer: Da 101.945; q_net 2; logP -8.9872[[ (H-2O-1H1) → (K1Na2) ]], Compomer: Da 111.911; q_net 2; logP -11.2898[[ (H-2O-1Na1) → (K3) ]], Compomer: Da 112.963; q_net 2; logP -9.90349[[ (H-2O-1H1) → (H4N1K2) ]], Compomer: Da 115.882; q_net 2; logP -7.82405[[ (H1) → (K3) ]], Compomer: Da 116.866; q_net 2; logP -11.2898[[ (H-2O-1H4N1) → (K3) ]], Compomer: Da 117.919; q_net 2; logP -9.90349[[ (H-2O-1H1) → (K2Na1) ]], Compomer: Da 133.893; q_net 2; logP -10.8198[[ (H-2O-1H1) → (K3) ]], ]
ERROR MetaboliteAdductDecharger 3:266 Return code: -1073740791
ERROR MetaboliteAdductDecharger 3:266 Execute failed: Failed to execute node MetaboliteAdductDecharger
This is the same inconclusive error that you had in the other node. The log actually looks ok. Can you maybe monitor your RAM usage to see if your computer runs out of RAM?
Or maybe use FileFilter to test your full workflow on a subset of your sample first.
My RAM is16 G.
a)I want to ask that the MetaboliteAdductDecharger node does work
hard . I change the parameter charge_max and charge_span_max.
b)I can’t find the datasets in OpenMS/turo
16GB RAM should be enough. It is really surprising, we never had this problem before.
a) To reduce the decharging further you should reduce the potential_adducts as well.
b) The data is also here: index - powered by h5ai v0.28.1 (https://larsjung.de/h5ai/)
Thanks, I will try it again.
I try it again based on the new parameter.
But I don’t know where occurs wrong.
Failing process stdout: [Adding neutral: ---------- Adduct -----------------, Charge: 0, Amount: 1, MassSingle: -18.0106, Formula: H-2O-1, log P: -1.38629, , MassExplainer table size: 8, Generating Masses with threshold: -4.15888 …, done, 0 of 222707 valid net charge compomer results did not pass the feature charge constraints, Inferring edges raised edge count from 222707 to 383555, Found 383555 putative edges (of 24827862) and avg hit-size of 0.580639]
ERROR MetaboliteAdductDecharger 0:265
Failing process stderr: [Compomer: Da -35.0371; q_net 0; logP -2.81341[[ (H4N1) → (H-2O-1H1) ]], Compomer: Da -18.0106; q_net 0; logP -1.38629[[ () → (H-2O-1) ]], Compomer: Da -17.0265; q_net 0; logP -1.42712[[ (H4N1) → (H1) ]], Compomer: Da -0.984016; q_net 0; logP -2.81341[[ (H1) → (H-2O-1H4N1) ]], Compomer: Da 0.984016; q_net 0; logP -2.81341[[ (H-2O-1H4N1) → (H1) ]], Compomer: Da 17.0265; q_net 0; logP -1.42712[[ (H1) → (H4N1) ]], Compomer: Da 18.0106; q_net 0; logP -1.38629[[ (H-2O-1) → () ]], Compomer: Da 35.0371; q_net 0; logP -2.81341[[ (H-2O-1H1) → (H4N1) ]], ]
ERROR MetaboliteAdductDecharger 0:265 Return code: -1073740791
I still would like to see the memory usage. Can you please track it? You are running an optimization problem on almost 400.000 entries here.
Yes, the error may be relation with it . How can I reduce them obviously?
a) Choose the fit auuucts.
b) Limit parameters.
You have to change the settings in FeatureFinderMetabo and make them a bit stricter.
E.g. require a higher chrom_peak_snr (e.g. 4) or noise_threshold_int (e.g. 20000).
Use a narrower mass_to_charge tolerance (e.g. lower to 10 or even 5 ppm).
A stricter isotope_filtering_model (2% RMSE), which types of compounds are you looking for anyway?
This is the RAM
First in KNIME
Second in window.
I want to ask the node is work fluently in window or linux.
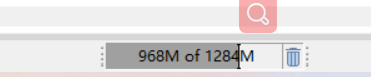

yes this looks fine. Is it getting higher or is this the maximum?
It is higher higher.
Now, there is an error as before.
And what was the maximum memory it used?
It is about 96%,but it isn’t stuck. The KNIME once exited possibly because of the limited RAM when I open the webpage.
Ok,I want to look for lipid compounds.And I am exploring the suitable method.
To be completely honest here, for lipidomics, I would suggest using a tool that was specifically designed for DIA analysis of lipids (e.g. MS-DIAL)
