Hi, all
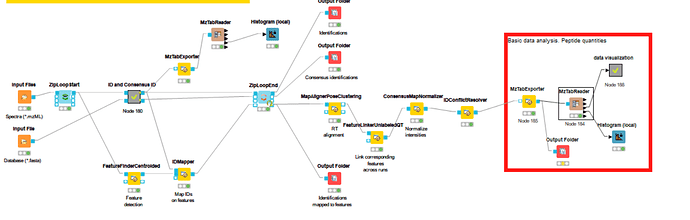
Now I have excuted the workflow. I have two question about this.
a) The FeatureFinderCentroided node is so slow, It continue more than 24h. I use three data to test the workflow.
b) The mztable exports two table(PSM and peptide), So which table should be considered. and does the feature intensity column exist the table?
Thank you!
Hi,
the runtime depends on too many factors:
- How big is your data
- How much memory/cores does your computer have
- What is the intensity threshold that you configured
Regarding mzTab, please look at the mzTab specification:
https://www.psidev.info/mztab
It depends on what you want to do.
Ok, thank you very much!
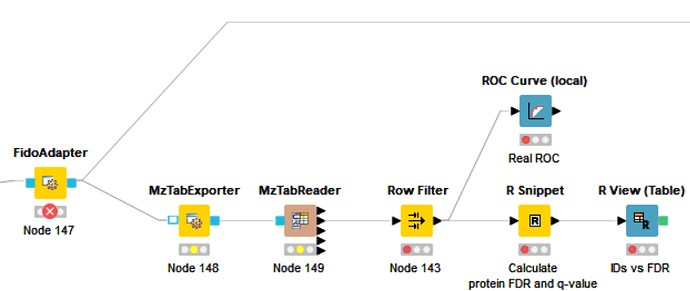
The new flow have another problem about the FidoAdapter.
Reading input data…
Generating temporary files for Fido…
Error: Unexpected internal error (Error: Unsuitable score type for peptide-spectrum matches detected (problem: lower scores are better).
Fido requires probabilities as scores, e.g. as produced by IDPosteriorErrorProbability with the ‘prob_correct’ option.)
The node parameter
Thanks
The important part are the nodes before FidoAdapter.
And which version of the OpenMS plugin are you using?
eh, it‘s 2.3 version.
Could you please try to update to 2.7?
Yeah, of course, I could.
I upgrad the version, but it is still the same wrong as before.
Error: Unsuitable score type for peptide-spectrum matches detected (problem: lower scores are better).
Fido requires probabilities as scores, e.g. as produced by IDPosteriorErrorProbability with the ‘prob_correct’ option.
Did you do this?
I can’t open the link. I just conduct a new workflow according the old flow.
Now, I enter the webpage. I am reading it.
The KNIME version is 4.6 and the OpenMS is 2.7.
This topic was automatically closed 90 days after the last reply. New replies are no longer allowed.