Hi, all
Is there about the workflow about proteomics?
I want to use the workflow to analysis the data.
Thank you very much!
Hi!
Yes, there should be some workflows on the KNIME hub:
Which are also described in the KNIME blog:
HTH
Julianus
Thank you very much!
Ok,Thank you! I will try it.
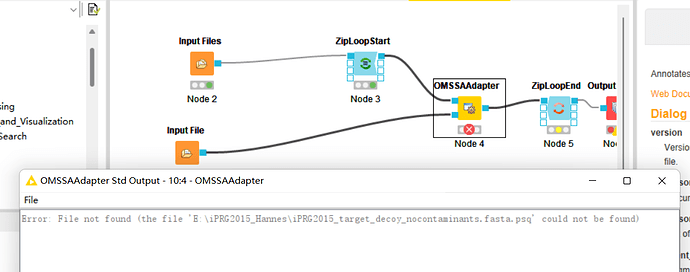
Error: File not found (the file ‘E:\iPRG2015_Hannes\iPRG2015_target_decoy_nocontaminants.fasta.psq’ could not be found)
I dowload the file,but it’s wrong.
I think you downloaded the wrong folder.
It is there: index - powered by h5ai v0.28.1 (https://larsjung.de/h5ai/)
Yes, I am wrong. I dowload the file to test the flow. Thank you!
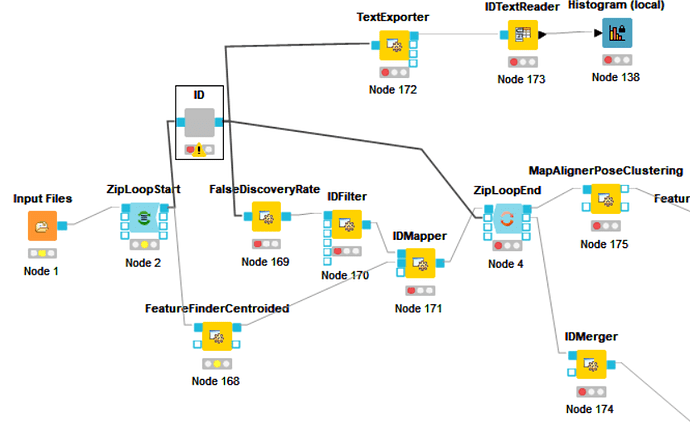
Yes, the workflow can work. The .fasta, .fasta.pin, .fasta.phr input the inputfile respecticely, Then The worlflow can be flow. So I have a conflusion that The InputFile can contain three files. followed the storage then is excuted. It’s magical!
@jpfeuffer The node ID should contain the nodes(Input files OMSSAAdapter peptideindexer). Is it right?
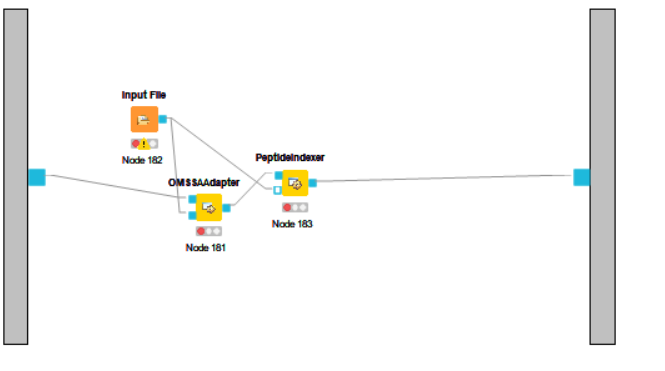
@jpfeuffer Hi, the flow occures the error.
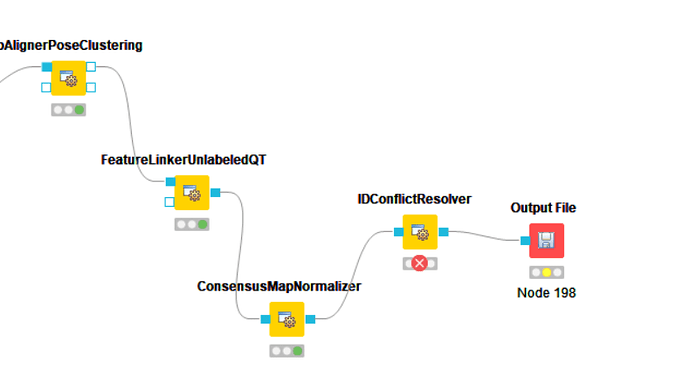
Error: Unable to write file (the file ‘F:\KNIME_temporary\knime_openmsLFQ_iPRG245011\fs-IDCon_11-197-45027\000\000\IDConflictResolver_0\JD_06232014_sample2_A.featureXML’ could not be created. invalid file extension, expected ‘consensusXML’)
Hi, please try to set the OutputType in the configuration dialogue of IDConflictResolver to consensusXML
Yeah,I got it!Thank you very much!
This topic was automatically closed 90 days after the last reply. New replies are no longer allowed.