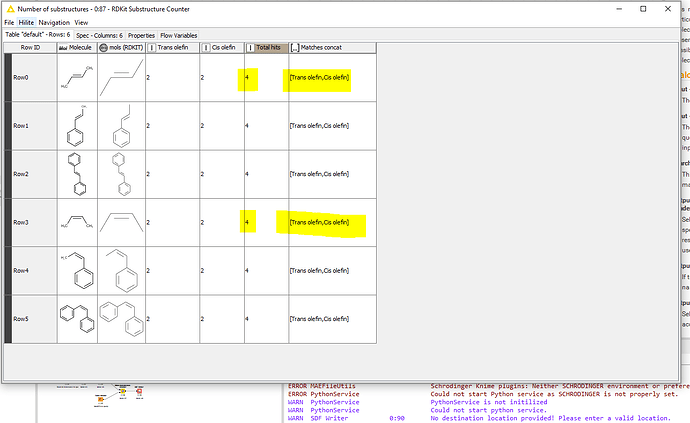
Hello! I’m trying to use the RDKit Substructure Counter to do substructure searches on an SD file, and I’d like to be able to distinguish cis vs trans olefins in my SMARTS queries. However, when I run the workflow, I get matches for cis and trans olefins with both my cis and trans SMARTS queries. I’ve attached a workflow and a screenshot.). Any help is appreciated, thanks!
Benji
SMARTS_substructure_test.knwf (27.3 KB)