Hi Knime community members,
I was wondering if anyone could help me with getting some traction in Knime for a fundus photo train/test project in exudative retinal diseases. I have tried loading nodes of fundus photos, trying to define their pathologic findings by Canny or Sobel algorithms or model photos and interrogating for statistical correlation in Knime. However, when I try to move from one phase to another I am not able to get the workflow or even any kind of pattern through Knime. I seem to get blocked.
I would be most appreciative of any assistance stepwise or in the large picture how to gain continuity and traction in this project.
Thanks for your interest,
Joseph Eichenbaum
Hi @J_Eichenbaum, welcome onboard!
Thanks for sharing your project — it sounds like a fascinating and meaningful application of KNIME.
If you’re comfortable, feel free to share:
- A sample fundus image or a small anonymized dataset
- A screenshot of the point where your workflow gets stuck
- A brief explanation of the labels you’re using and what kind of output you’re aiming for
That would really help the community provide more targeted support — or even suggest example workflows that are closer to your goal.
You’re clearly working on something impactful, and once the pipeline starts flowing, KNIME can really shine with medical image processing.
Wishing you the best of luck — and happy to help if you share a bit more detail!
Warm regards,
Alpay
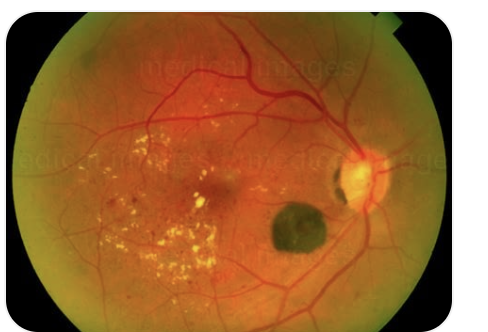
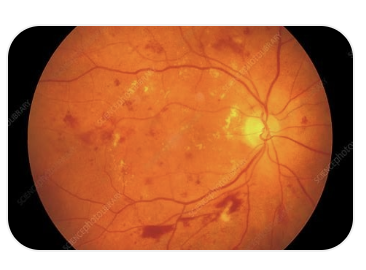
Hi Alpay,
I have uploaded 2 sample fundus photos with Diabetic retinopathy, obtained from my desktop; however, when I try to access fundus photos that I have stored in Local or via Explorer, I get “show warning dialog when connecting to server” message under Prompt.
I have not placed labels yet; I retain the fundus photos in a desktop file with e.g. Diabetic Retinopathy, Macula Degeneration etc; I would rather not label at the early stage and let the pathology fall into place by virtue of the Knime photo interrogation and edge characteristics of fundus findings. The desired output is to be able to take a fundus photo with the consent of the patient and check if the photo carries any of the diverse elements of ocular pathology, say diabetes, hypertension or sarcoid etc.or is in the range of normals. The latter will be a challenge because the range of normal has to be age adjusted; but with enough fundus image data, that range of normal can be obtained say by decade and with connecting database software we could store it by the decade.
Thanks so much for your interest.
Regards,
Joseph Eichenbaum
Hi @J_Eichenbaum.
It seems to me that your project is realy of great value. The ultimate goal is to be able to identify in a new retinal fundus image the probability of similarity with a disease previously learned by the retinal fundus model.
I’m not an expert, but taking the playful difference out of the equation, it bears a strong resemblance to a much-reviewed problem of identifying cats and dogs in photos aproach, but this time with deseases.
I found this links for you.
Take a look
Cats and dogs
Skin cancer
Plants
Its a start to learn using knime for your goal.
Best regards
Hi HMFA,
Many thanks for your thoughtful reply.
I will look into these.
Regards,
Joseph Eichenbaum
This topic was automatically closed 90 days after the last reply. New replies are no longer allowed.