Hi,
I’m getting a “execute failed:null pointer exception: Null” error with the PMI node. I’ve generated 3D coordinates using RDKIT, and also tried it directly on the SDF (2D coordinates only), but to no avail. I’m running 4.1.3 on a Mac (Catalina).
Thanks,
Natasja
Hi @natasja, it would be helpful if you could provide the sd file with the molecules you’re trying to analyze, as well as the workflow you’re using.
Hi Natasja,
Thanks for reporting this to us. Would you be able to look in either the KNME console or log view (Goto the View menu and select ‘Open KNIME Log’ and copythe additional information in there following the NullPointerException?
It should look something vaguely like the following (but not exactly - I forced this to happen in a Java Snippet node deliberately just now!):
java.lang.NullPointerException
at JSnippet.snippet(JSnippet.java:25)
at org.knime.base.node.jsnippet.JavaSnippetCellFactory.getCells(JavaSnippetCellFactory.java:300)
at org.knime.base.node.jsnippet.JavaSnippet.execute(JavaSnippet.java:1023)
at org.knime.base.node.jsnippet.JavaSnippetNodeModel.execute(JavaSnippetNodeModel.java:139)
at org.knime.core.node.NodeModel.execute(NodeModel.java:737)
at org.knime.core.node.NodeModel.executeModel(NodeModel.java:571)
at org.knime.core.node.Node.invokeFullyNodeModelExecute(Node.java:1236)
at org.knime.core.node.Node.execute(Node.java:1016)
at org.knime.core.node.workflow.NativeNodeContainer.performExecuteNode(NativeNodeContainer.java:557)
at org.knime.core.node.exec.LocalNodeExecutionJob.mainExecute(LocalNodeExecutionJob.java:95)
at org.knime.core.node.workflow.NodeExecutionJob.internalRun(NodeExecutionJob.java:218)
at org.knime.core.node.workflow.NodeExecutionJob.run(NodeExecutionJob.java:124)
at org.knime.core.util.ThreadUtils$RunnableWithContextImpl.runWithContext(ThreadUtils.java:334)
at org.knime.core.util.ThreadUtils$RunnableWithContext.run(ThreadUtils.java:210)
at java.util.concurrent.Executors$RunnableAdapter.call(Executors.java:511)
at java.util.concurrent.FutureTask.run(FutureTask.java:266)
at org.knime.core.util.ThreadPool$MyFuture.run(ThreadPool.java:123)
at org.knime.core.util.ThreadPool$Worker.run(ThreadPool.java:246)
Thanks
Steve
(@Vernalis’ alter-ego)
Hi Steve,
Here you go:
2020-07-01 15:07:47,742 : ERROR : KNIME-Worker-541-Principal Moment of Intertia (PMI)-Derived Properties 3:58 : : Node : Principal Moment of Intertia (PMI)-Derived Properties : 3:58 : Execute failed: (“NullPointerException”): null
java.lang.NullPointerException
Thanks,
Natasja
More or less figured it out I think. It doesn’t like the “enhanced” chiral annotations:
I confirmed this with 99% confidence. I used the speedy chiral smiles filter (thank you Vernalis’ alter ego and coworkers!) filter on a ~30K compound set that was failing before. After removal of the ~1000 chiral compounds, it runs just fine. Fails on the set with ~1000 chiral compounds.
Natasja
Hi Natasja,
Thanks for that - that really helps to see where the problem might be coming from. (And glad you like the Speedy SMILES node  ) Would you be able to copy/paste the SDF table cell for that molecule into a reply so that I can use it to track down the problem?
) Would you be able to copy/paste the SDF table cell for that molecule into a reply so that I can use it to track down the problem?
Thanks
Steve
Here you go:
Mrv15a5 10211913272D
0 0 0 0 0 999 V3000
M V30 BEGIN CTAB
M V30 COUNTS 20 21 0 0 0
M V30 BEGIN ATOM
M V30 1 C 6.1045 -7.4935 0 0
M V30 2 N 6.1045 -5.9535 0 0
M V30 3 C 4.7708 -5.1835 0 0
M V30 4 C 3.4374 -5.9535 0 0
M V30 5 O 3.4374 -7.4935 0 0
M V30 6 N 2.1037 -5.1835 0 0
M V30 7 C 0.77 -5.9535 0 0 CFG=2
M V30 8 C 0 -7.2872 0 0
M V30 9 C -0.77 -5.9535 0 0 CFG=1
M V30 10 C -2.1037 -5.1835 0 0
M V30 11 C -3.4374 -5.9535 0 0
M V30 12 C -4.7708 -5.1835 0 0
M V30 13 C -4.7708 -3.6437 0 0
M V30 14 C -3.4374 -2.8737 0 0
M V30 15 C -2.1037 -3.6437 0 0
M V30 16 Cl -0.77 -2.8737 0 0
M V30 17 C 7.4382 -5.1835 0 0
M V30 18 O 7.4382 -3.6437 0 0
M V30 19 C 8.7719 -5.9535 0 0
M V30 20 C 10.1056 -5.1835 0 0
M V30 END ATOM
M V30 BEGIN BOND
M V30 1 1 1 2
M V30 2 1 2 3
M V30 3 1 3 4
M V30 4 2 4 5
M V30 5 1 4 6
M V30 6 1 7 6 CFG=3
M V30 7 1 7 8
M V30 8 1 8 9
M V30 9 1 7 9
M V30 10 1 9 10 CFG=1
M V30 11 1 10 11
M V30 12 2 11 12
M V30 13 1 12 13
M V30 14 2 13 14
M V30 15 1 14 15
M V30 16 2 10 15
M V30 17 1 15 16
M V30 18 1 2 17
M V30 19 2 17 18
M V30 20 1 17 19
M V30 21 2 19 20
M V30 END BOND
M V30 BEGIN COLLECTION
M V30 MDLV30/STERAC1 ATOMS=(2 7 9)
M V30 END COLLECTION
M V30 END CTAB
M END
292.76
Z2715607261
4.6
1
2
<Rot_bonds>
5
2
1.63646
49.41
0.333333
$$$$
1 Like
Thanks @natasja - I will look into this tomorrow.
Steve
Hi Natasja,
I have just put your structure (just with 2D coordinates) into a very simple workflow:
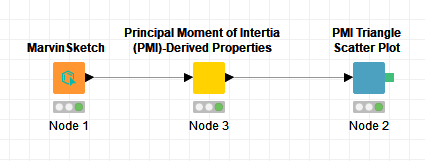
And the output is expected:
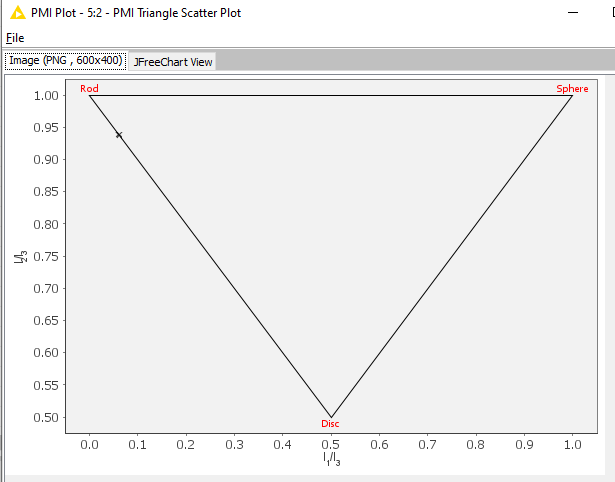
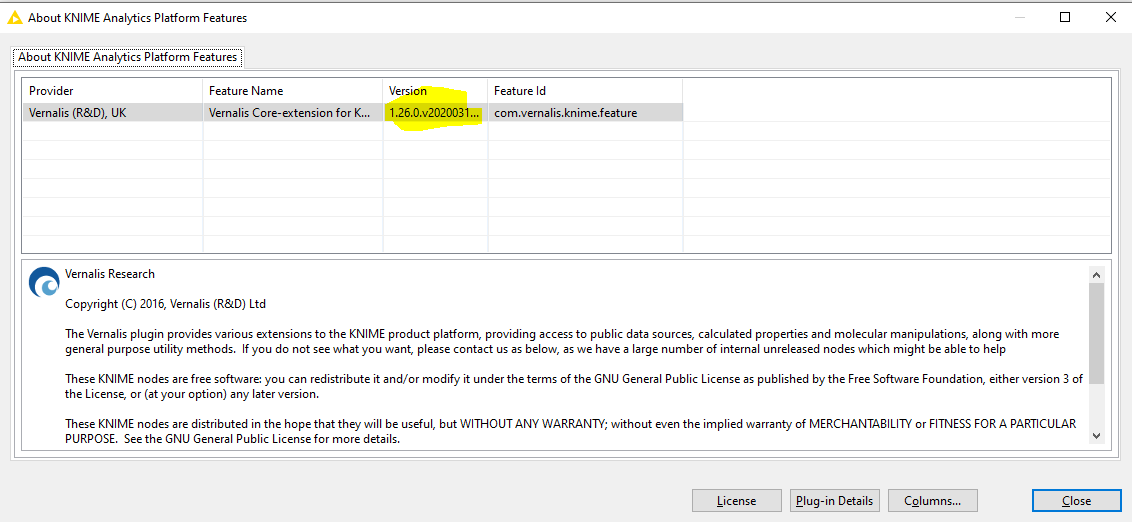
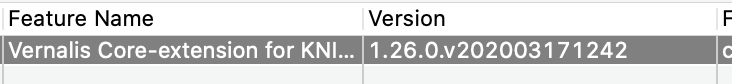
Could I check, is this the versions of the nodes you are using? Would you be able to check which version of the Vernalis nodes you currently have installed? (You can do this from the ‘Help’ menu - click on 'About KNIME analytics platform, then on the Vernalis logo:

You should get a dialog something like this:
If you could let me know the version in the highlighted box that would be helpful.
Yours,
Steve
Thanks Natasja. Could you also clarify which node is failing?
Steve
OK, I’m still not managing to reproduce this at the moment (but, I dont have access to a Mac, although I dont imagine that’s a problem!). Let’s keep going!
Could you ensure that your KNIME desktop console is set to ‘DEBUG’ - goto ‘Preferences’ in the ‘File’ menu and browse to KNIME GUI:
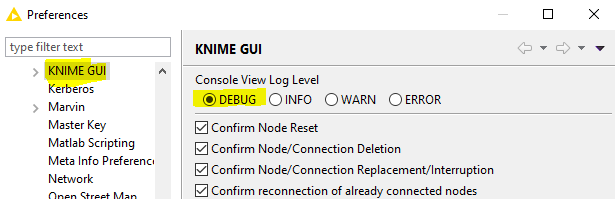
Then, re-run again, and when the node crashes, if you could include all the information in the lines which follow the line with java.lang.NullPointerException (they should all be indented and start with 'at ’ as in one of the earlier responses above) - hopefully that will at least show me exactly where things are going wrong in the code!
Steve
CONSOLE:
0 …" (index 0) in row “Row0” is SDF and does not comply with its supposed superclass RDKit Molecule
WARN Principal Moment of Intertia (PMI)-Derived Properties 0:272 The selected column (Molecule (with coord.)) is not present in the incoming table
ERROR Principal Moment of Intertia (PMI)-Derived Properties 0:272 Execute failed: (“NullPointerException”): null
KNIME.log:
java.lang.NullPointerException
at com.vernalis.knime.chem.util.points.AbstractPoint.getCentreOfGravity(AbstractPoint.java:287)
at com.vernalis.knime.chem.pmi.props.PMI.fromPoints(PMI.java:67)
at com.vernalis.knime.chem.pmi.nodes.props.pmiderived.PmiDerivedPropsNodeFactory$1.getObjFromPointList(PmiDerivedPropsNodeFactory.java:58)
at com.vernalis.knime.chem.pmi.nodes.props.pmiderived.PmiDerivedPropsNodeFactory$1.getObjFromPointList(PmiDerivedPropsNodeFactory.java:1)
at com.vernalis.knime.core.nodes.props.abstrct.AbstractPointListMoleculePropertyCalcNodeModel.getMolContainerFromCell(AbstractPointListMoleculePropertyCalcNodeModel.java:128)
at com.vernalis.knime.core.nodes.props.abstrct.AbstractMoleculePropertyCalcNodeModel.getContainerObjFromCell(AbstractMoleculePropertyCalcNodeModel.java:175)
at com.vernalis.knime.nodes.propcalc.AbstractPropertyCalcNodeModel$1.getCells(AbstractPropertyCalcNodeModel.java:222)
at org.knime.core.data.container.RearrangeColumnsTable.calcNewCellsForRow(RearrangeColumnsTable.java:541)
at org.knime.core.data.container.RearrangeColumnsTable.calcNewColsSynchronously(RearrangeColumnsTable.java:462)
at org.knime.core.data.container.RearrangeColumnsTable.create(RearrangeColumnsTable.java:380)
at org.knime.core.node.ExecutionContext.createColumnRearrangeTable(ExecutionContext.java:385)
at org.knime.core.node.streamable.simple.SimpleStreamableFunctionNodeModel.execute(SimpleStreamableFunctionNodeModel.java:117)
at org.knime.core.node.NodeModel.execute(NodeModel.java:737)
at org.knime.core.node.NodeModel.executeModel(NodeModel.java:571)
at org.knime.core.node.Node.invokeFullyNodeModelExecute(Node.java:1236)
at org.knime.core.node.Node.execute(Node.java:1016)
at org.knime.core.node.workflow.NativeNodeContainer.performExecuteNode(NativeNodeContainer.java:557)
at org.knime.core.node.exec.LocalNodeExecutionJob.mainExecute(LocalNodeExecutionJob.java:95)
at org.knime.core.node.workflow.NodeExecutionJob.internalRun(NodeExecutionJob.java:218)
at org.knime.core.node.workflow.NodeExecutionJob.run(NodeExecutionJob.java:124)
at org.knime.core.util.ThreadUtils$RunnableWithContextImpl.runWithContext(ThreadUtils.java:334)
at org.knime.core.util.ThreadUtils$RunnableWithContext.run(ThreadUtils.java:210)
at java.util.concurrent.Executors$RunnableAdapter.call(Executors.java:511)
at java.util.concurrent.FutureTask.run(FutureTask.java:266)
at org.knime.core.util.ThreadPool$MyFuture.run(ThreadPool.java:123)
at org.knime.core.util.ThreadPool$Worker.run(ThreadPool.java:246)
2020-07-06 11:25:22,281 : DEBUG : KNIME-Worker-272-Principal Moment of Intertia (PMI)-Derived Properties 0:272 : : WorkflowManager : Principal Moment of Intertia (PMI)-Derived Properties : 0:272 : Principal Moment of Intertia (PMI)-Derived Properties 0:272 doBeforePostExecution
2020-07-06 11:25:22,282 : DEBUG : KNIME-Worker-272-Principal Moment of Intertia (PMI)-Derived Properties 0:272 : : NodeContainer : Principal Moment of Intertia (PMI)-Derived Properties : 0:272 : Principal Moment of Intertia (PMI)-Derived Properties 0:272 has new state: POSTEXECUTE
2020-07-06 11:25:22,282 : DEBUG : KNIME-Worker-272-Principal Moment of Intertia (PMI)-Derived Properties 0:272 : : WorkflowManager : Principal Moment of Intertia (PMI)-Derived Properties : 0:272 : Principal Moment of Intertia (PMI)-Derived Properties 0:272 doAfterExecute - failure
2020-07-06 11:25:22,282 : DEBUG : KNIME-Worker-272-Principal Moment of Intertia (PMI)-Derived Properties 0:272 : : Node : Principal Moment of Intertia (PMI)-Derived Properties : 0:272 : reset
2020-07-06 11:25:22,282 : DEBUG : KNIME-Worker-272-Principal Moment of Intertia (PMI)-Derived Properties 0:272 : : Node : Principal Moment of Intertia (PMI)-Derived Properties : 0:272 : clean output ports.
2020-07-06 11:25:22,282 : DEBUG : KNIME-Worker-272-Principal Moment of Intertia (PMI)-Derived Properties 0:272 : : WorkflowDataRepository : Principal Moment of Intertia (PMI)-Derived Properties : 0:272 : Removing handler 70357fb6-bd5d-4778-9759-7af79713ce4f (Principal Moment of Intertia (PMI)-Derived Properties 0:272: ) - 153 remaining
2020-07-06 11:25:22,282 : DEBUG : KNIME-Worker-272-Principal Moment of Intertia (PMI)-Derived Properties 0:272 : : NodeContainer : Principal Moment of Intertia (PMI)-Derived Properties : 0:272 : Principal Moment of Intertia (PMI)-Derived Properties 0:272 has new state: IDLE
2020-07-06 11:25:22,282 : DEBUG : KNIME-Worker-272-Principal Moment of Intertia (PMI)-Derived Properties 0:272 : : Node : Principal Moment of Intertia (PMI)-Derived Properties : 0:272 : Configure succeeded. (Principal Moment of Intertia (PMI)-Derived Properties)
2020-07-06 11:25:22,282 : DEBUG : KNIME-Worker-272-Principal Moment of Intertia (PMI)-Derived Properties 0:272 : : NodeContainer : Principal Moment of Intertia (PMI)-Derived Properties : 0:272 : Principal Moment of Intertia (PMI)-Derived Properties 0:272 has new state: CONFIGURED
Thank you - that’s very helpful as it let’s me see where the code is breaking. At the moment, I still can’t see eactly why that is happening but I’m beginning to get a vague idea. Would you be able to do 2 things?
- Could you download this workflow (72.7 KB) into KNIME - it should be pre-executed, and then try re-setting and re-running ‘Node 3’, followed by ‘Node 5’ and see if they both work. If those both work OK, could you then try resetting ‘Node 4’ and re-running ‘Node 3’, and then resetting ‘Node 1’ and re-running all the nodes?
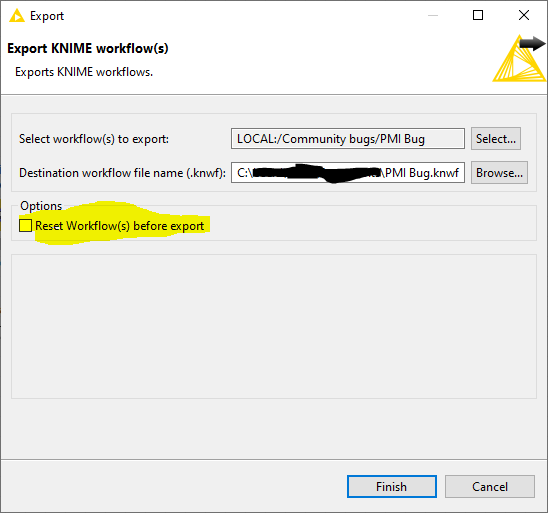
- If you could export a workflow contaning one of your failing molecules (the one in the posts above would be good), without resetting during export, and attach it to a reply? (I want to see if there are any subtle differences in the V3000 mol block written by the version of Marvin you are using)
Thanks again, and sorry this it taking to long to resolve,
Steve
Hi Steve,
For the delay, but yes, both nodes 3 & 5 work fine after the reset. I also copied the node into my workflow with the larger SDF file and there it still fails. Not sure if the copy/paste made additional changes in the node?
Thanks,
Natasja
Thanks @natasja. Copying the node over shouldnt make any difference, so it looks like the problem is something to do with exactly how the SDF file is formatted.
Could you use this ‘component’ in your workflow at the same place as the node which is breaking?
You will need to select ‘configure’ on this node, and change the dropdown to the sd-file column in your incoming table:
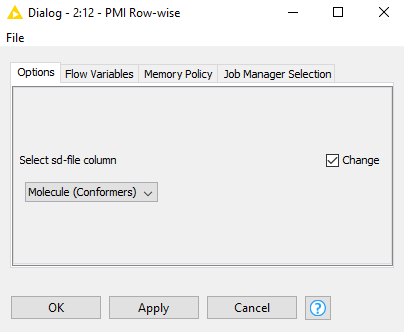
It should run until it reaches a failing row. The sd-file of that row will be saved in your knime workspace in the folder containing the workflow (if you don’t know how to get to that, please let me know and I will show you). Please attach that file back in a reply.
Thanks
Steve
The uploaded SDF file was zipped using gzip, but I forced it to use .zip as the extension for the upload. Also pasted below.
Thanks,
Natasja
Here you go:
RDKit 3D
0 0 0 0 0 0 0 0 0 0999 V3000
M V30 BEGIN CTAB
M V30 COUNTS 20 21 0 0 0
M V30 BEGIN ATOM
M V30 1 C -3.86734 -1.38349 1.14107 0
M V30 2 N -3.56667 -0.952971 -0.200643 0
M V30 3 C -2.82218 -1.81529 -1.04437 0
M V30 4 C -1.36547 -1.65013 -0.993184 0
M V30 5 O -0.690791 -2.40813 -1.73623 0
M V30 6 N -0.636319 -0.74452 -0.21143 0
M V30 7 C 0.946231 -0.80371 -0.375045 0 CFG=2
M V30 8 C 1.216 -1.18611 1.29601 0
M V30 9 C 1.25373 0.449202 0.796162 0 CFG=1
M V30 10 C 2.82716 0.574488 0.538654 0
M V30 11 C 3.65175 -0.467995 0.72975 0
M V30 12 C 5.01952 -0.391817 0.514819 0
M V30 13 C 5.49978 0.830138 0.0872186 0
M V30 14 C 4.63633 1.89691 -0.105561 0
M V30 15 C 3.29687 1.78464 0.116017 0
M V30 16 Cl 2.18109 3.09803 -0.113906 0
M V30 17 C -3.99839 0.292286 -0.669175 0
M V30 18 O -3.72765 0.661529 -1.85376 0
M V30 19 C -4.76754 1.26316 0.0997814 0
M V30 20 C -5.08612 0.953777 1.33291 0
M V30 END ATOM
M V30 BEGIN BOND
M V30 1 1 1 2
M V30 2 1 2 3
M V30 3 1 3 4
M V30 4 2 4 5
M V30 5 1 4 6
M V30 6 1 7 6 CFG=1
M V30 7 1 7 8
M V30 8 1 8 9
M V30 9 1 7 9
M V30 10 1 9 10 CFG=3
M V30 11 2 10 11
M V30 12 1 11 12
M V30 13 2 12 13
M V30 14 1 13 14
M V30 15 2 14 15
M V30 16 1 10 15
M V30 17 1 15 16
M V30 18 1 2 17
M V30 19 2 17 18
M V30 20 1 17 19
M V30 21 2 19 20
M V30 END BOND
M V30 BEGIN COLLECTION
M V30 MDLV30/STERAC1 ATOMS=(2 7 9)
M V30 END COLLECTION
M V30 END CTAB
M END
292.76
Z2715607261
Enamine32K
1.9564999999999997
49.410000000000004
292.097855464
5
1
2
2
<SMILES (Canonical)>
C=CC(=O)N(C)CC(=O)N[C@@H]1C[C@H]1c1ccccc1Cl
$$$$broken.sdf.zip (890 Bytes)
Thanks - that’s very helpful, as I can also reproduce the error using that file. I will get back to you when I have figured out why it breaks!
Steve
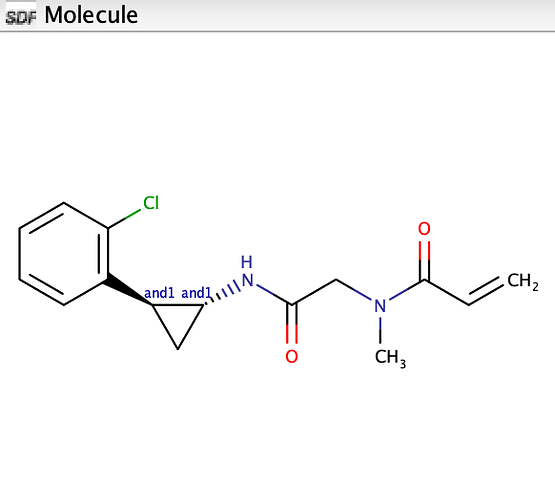
 ) Would you be able to copy/paste the SDF table cell for that molecule into a reply so that I can use it to track down the problem?
) Would you be able to copy/paste the SDF table cell for that molecule into a reply so that I can use it to track down the problem?