Hi,
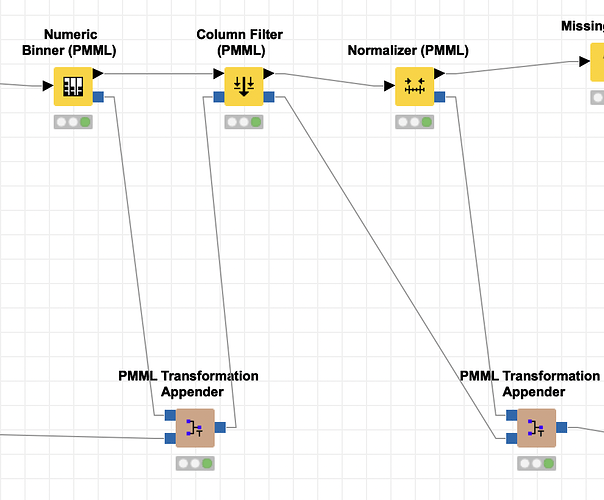
Thanks @mlauber71. I had a look at the workflow and I think I know what the main problem is: the Column Filter (PMML) filters the Data Dictionary of an incoming PMML document and outputs that, but PMML does not have any specification for filtering columns. So the new PMML created by the node does not really document the removal of columns, it just deletes every mention of them from the spec of the input data (the DataDictionary). It also keeps the TransformationDictionary as-is, so now the binner spec from the previous node comes into the PMML Transformation Appender from the top and from the bottom. The Appender now thinks that a new transformation for age_binned has been added and adds another entry in the TransformationDictionary, which is “overwriting” the old one, without really doing something new because it is actually just a copy. The Column Filter (PMML) must therefore be connected like in the screenshot below.
This issue is kind of a weird result of PMML not supporting column filtering/replacement and KNIME not documenting the usage of the PMML nodes properly here.
The Gradient Boosted Trees Model, by the way, is not supported by the PMML Compiler. The following model types are supported:
- Regression
- SVM
- MLP
- K-Means Clustering
- Decision Trees
- Naive Bayes
- Rulesets
- Ensembles of the above (but not boosting)
Kind regards,
Alex