@TheLeo
Thanks for the data, that helps a bit. Three thoughts/ideas for you to consider (this will be a long post as I will include examples).
I’ve uploaded the workflow that contains all the examples to KNIME hub here.
1/ Convert data/time to ISO8601 string.
When writing dates and times to file formats/ databases that don’t support a native date/time type, then ISO8601 format strings should be used. This is an internationally recognised standard and widely supported across software platforms/packages. When reading an ISO8601 string Pandas can be configured to parse the string into a Pandas datetime dtype.
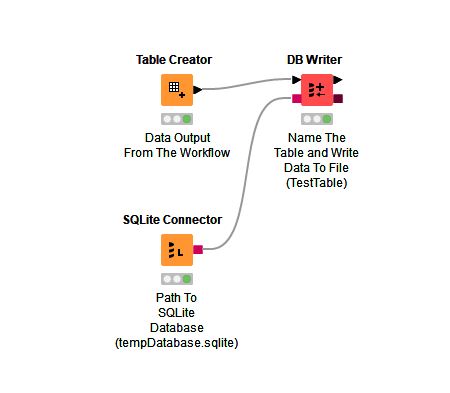
In KNIME you would convert the dateTime column to a string.
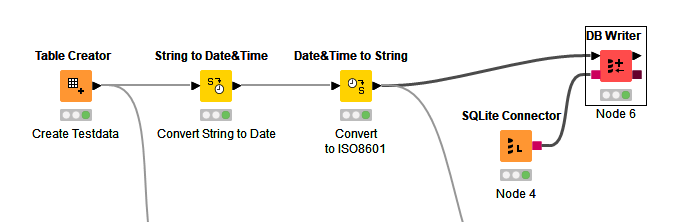
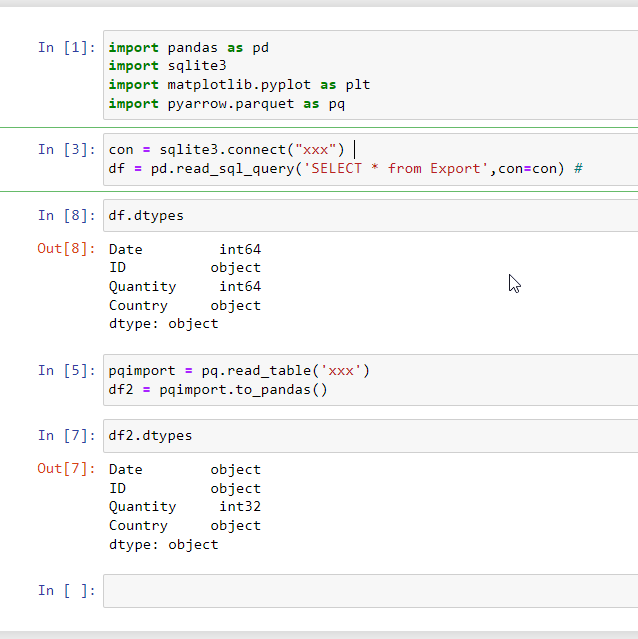
And in Python you would configure Pandas to parse the string. The additional parameter compared with my prior post is parse_dates which is passed a list of columns containing ISO8601 format strings.
import sqlalchemy as db
import pandas as pd
engine = db.create_engine("sqlite:///D:\\data\\sqlite.sqlite")
with engine.connect() as db_connection:
table_name = 'Export'
df = pd.read_sql(f'SELECT * FROM {table_name}',
con=db_connection,
parse_dates=["Date"])
2/ Implement a data schema
Your country column is a categorical column and has a lot of redundant duplicate strings. This is going to increase your output file size considerably. You might want to consider creating dimension tables holding which are referenced by the original (fact) table. In KNIME this is relatively easy to do.
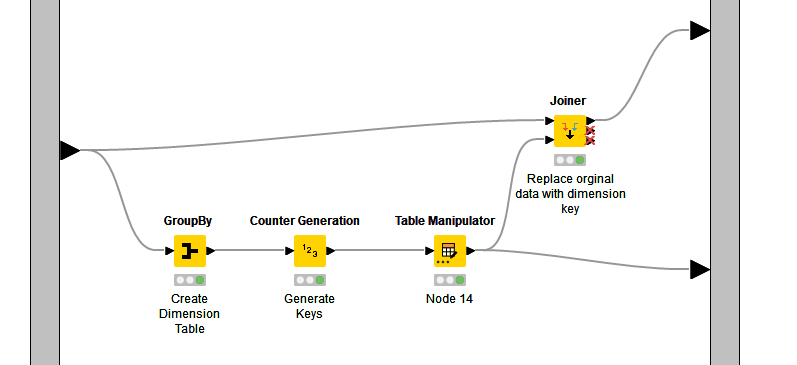
In the following workflow, the a dimension table is created by grouping the Country column in Create Testdata. A counter generator is used to generate a set of sequential keys (integers). The table is then manipulated to rename the counter generated int column to CountryKey. This table is then output from the metanode as the dimension table (bottom leg of the workflow).
The dimension table is then joined to the original table matching on the country column. The original Country column is dropped and the CountryKey is added. This replaces the duplicate Country strings with an integer which references the actual country name in the dimension table. This will reduce the size of your output file significantly.
The benefit of this approach is (a) it is good practice if you move to an online database; (b) it will help pickup errors in categorical fields if there are slightly misspelled items, missing items and other corruption. The downside is that the end user will need to use dimension tables to add the string representation to the fact/data file (though with Pandas this should not be too difficult).
3/ Consider HDF5 file format
HDF5 file format provides a greater degree of flexibility compared with many other file formats. The downside is that KNIME does not have a native node supporting it (I’ve never understood why not). It is possible to use a Python node to export data to an HDF5 file though it will require a bit of work due to the evolving nature of KNIME’s Python implementation.
Pre-requisites:
You will need a conda environment with the KNIME base packages, into which you will also need to add pytables. Note: There is another Python package h5py which provides more control, but for the purposes of keeping it simple I will stick with pytables.
Why Python in KNIME is a pain point
Pandas is a wrapper around underlying data types. This gives it great flexibility in abstracting functionality from implementation. When an operation is defined using pandas then its implementation is passed to the underlying data type to implement. When a third party package consumes a Pandas dataframe (such as Pytables) it assumes data is represented using convention data types such as int for integers and object for strings and Python datetime. However, KNIME does not conform to the conventions and so it stores strings as string instead of object and datatime as a java inspired something. It’s a pain!
The reason for the above rantorial is to explain why the following is more complicated than it needs to be.
Before adding the Python node, the date column needs to be converted to an ISO8601 string.
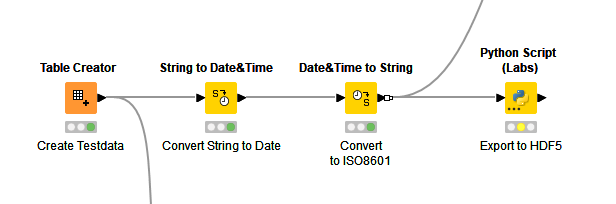
The Python script is as follows:
- Set the output file path and name of the table in the hdf5 file (hdf5 files can contain multiple tables in a folder like structure).
- Convert the ISO8601 date to Pandas datetime format, and convert strings to objects. Copy remaining columns to output file.
- Write output file setting compression mode and compression level.
import knime_io as knio
import pandas as pd
from os import path
# Configuration
output_path = path.abspath("D:\\Data\\dataExample.hdf")
output_table_name = "Export"
input_table = knio.input_tables[0].to_pandas()
output_table = pd.DataFrame()
# Convert ISO8601 dates to DateTime format.
dates_to_convert = ["Date"]
for column in dates_to_convert:
output_table[column] = pd.to_datetime(input_table[column],
format="%Y-%m-%d")
# Convert strings to objects
strings_to_convert = ["ID", "Country"]
for column in strings_to_convert:
output_table[column] = input_table[column].astype(object)
# Copy remaining columns to output table.
all_other_columns = ["Quantity"]
for column in all_other_columns:
output_table[column] = input_table[column]
# Output to hdf file
# mode = write a new file (w)
# complib = compression library
# complevel = compression level (1=faster, 9=more compression).
output_table.to_hdf(output_path,
output_table_name,
mode="w",
complib="zlib",
complevel=5)
# Pass though input.
knio.output_tables[0] = knio.write_table(input_table)
In Python to read the file you would use the following:
import pandas as pd
from os import path
input_path = path.abspath("D:\\Data\\dataExample.hdf")
input_table_name = "Export"
df = pd.read_hdf(input_path, key=input_table_name )
I hope this helps.