@helfortuny this sounds like an interesting study. These things come to my mind:
You indeed have a lot of variables that will need some preprocessing. One thing you could try is read about vtreat [you can find KNIME workflows for that here] and try that with your target and see what recommendations would be made to see if it makes any difference. You might have tried and removed highly correlated variables. At the end of the vtreat article there are links to other dimension reduction techniques you might want to explore, namely PCA. This would reduce your numbers but also obscure what is in such a component.
How is the quality of your data. Since it is specifically generated I assume the quality is quite good, but you should check for columns with many missings.
Then: out of the 300 cases how many are ‘positiv’? How unbalanced is your dataset? If it is very unbalanced you might find it difficult to get good results. In any case you might want to use AUCPR or LogLoss as your evaluation metric to focus on your true cases (XGBoost Predictor - #5 by mlauber71).
Your descriptive variables (age …) might not be used because of the massive amount of other data. You could try target encoding and then normalisation if you expect these features to have a permanent influence and your sample might well represent your future patients.
Then about normalisation: a simple transformation into 0-1 might not be sufficient if you have a very wide range of values, smaller differences in numbers in a lot of cases might get obscured by a few very large values. You could try and use logarithm to keep the ‘shape’ (and rank) but lift the lower numbers up or decimal scaling (available in the KNIME node).
Concerning models: For a lot of cases I prefer to use robust tree models (GBM, XGBoost, LightGBM) and boosters and auto-machine learning approaches. For such large numerical tasks if you cannot find a good approach there might be a case to try deep-learning.
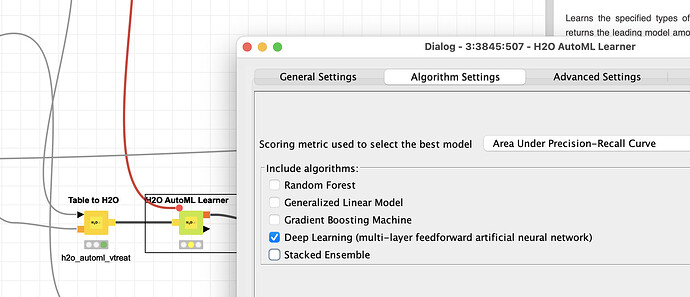
One way to determine if deep-learning might be able to help would be to use the H2O.ai AutoML node and just let it run deep-learning approaches in one branch to see if this can beat your other approaches.
Since this is a KNIME forum you might want to explore its opportunities. DL models will present a challenge of their own - starting with finding a good model architecture. But there is a book that might be worth reading - you can also search for videos from the authors.