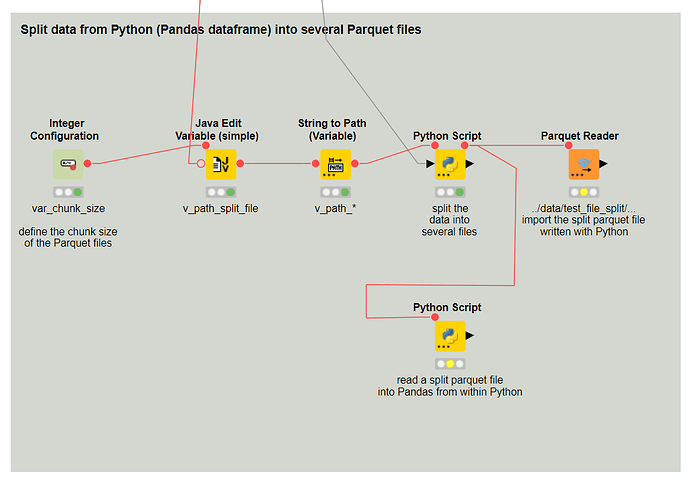
Carsten,
Thank you for reaching out!
Thrilled to hear you are working on this for 5.3 When is that due to be out?
Also…splitting the file does not feel like a great option anyway.
I am using pandas.
…here is the script:
“”"
This script will merge the results of a random list of series with the client transaction file.
Provide an input of the transaction file from the client, the list of series you would like to use
The script will access FRED, calculate change metrics, then merge the results into the transaction file, and then return the merged results back into the workflow.
After this merge, you will have to re-map these results to the output mask.
INSTRUCTIONS
- Supply the FRED Series names and plain-english names in the dict_series_list using a dictionary format
- Run the script
- Remember to assign the newly merged series to the output mask.
“”"
#Clear memory for script run
globals().clear()
Import requisite libraries
import pandas as pd
import numpy as np
import pyfredapi as pf
import os
import datetime
import time
Set runmode (1-Development in Spyder , 0 - Production KNIME)
runmode = 1
EXECUTION PARAMETERS
dict_series_list = {‘CPIRECSL’:‘CPI - Recreation’,
‘PCUATRNWRATRNWR’:‘PPI - Transp. & Warehousing’,
‘FEDFUNDS’:‘Federal Funds Effective Rate’,
‘USACPALTT01CTGYM’:‘CPI-ALL’,
‘wpu80’:‘Construction (Partial)’,
‘HOUST1F’:‘New Private Housing’,
‘RHEACBW027SBOG’:‘Real Estate Loans: Residential Real Estate Loans: Revolving Home Equity Loans, All Commercial Banks’
}
#input_path = “x”
input_path = “x”
#output_path = x"
output_path = “x”
Pull in transaction data file, import matplotlib if running in DEVELOPMENT mode
if runmode==1:
pd.set_option(‘display.max_columns’, None) # See all columns in pd dataFrame
pd.set_option(‘display.max_colwidth’, 20) # Limit column width in pd dataFrame
pd.set_option(‘display.expand_frame_repr’, False) # Remove truncated outputs
st_df = pd.read_csv(input_path+“InitialAssembly100_PCT.csv”, nrows=10000)
#st_df = pd.read_csv(input_path+“InitialAssembly100_PCT.csv”, nrows=10000)
import matplotlib.pyplot as plt
else:
import knime.scripting.io as knio # Imports KNIME libraries when in the KNIME environment
st_df = knio.input_tables[0].to_pandas()
Set FRED_API_KEY as an environmental variable
os.environ[‘FRED_API_KEY’] = ‘1c6a58b07dfeb3b40c85c4de15d4a84d’
Create a dictionary to store dataframes for each item
item_series_dfs = {}
def get_series(item):
#Pull series information and store it as a pandas dataframe
Series_df = pd.DataFrame(pf.get_series(series_id=item))
# Keep the date and value fields
Series_df = Series_df.iloc[:,-2:]
Series_df[item+'Series'] = item
Series_df[item+'SeriesName'] = dict_series_list[item]
# Format the date and extract Year/Month fields for merge with transaction data
Series_df['date'] = pd.to_datetime(Series_df['date'])
Series_df['Month'] = Series_df['date'].dt.month
Series_df['Year'] = Series_df['date'].dt.year
Series_df[item+'value'] = Series_df['value']
# Calculate 3-month and 12-month rolling averages
Series_df[item+'3MRoll'] = Series_df[item+'value'].rolling(window=3).mean()
Series_df[item+'12MRoll'] = Series_df[item+'value'].rolling(window=12).mean()
# Shift data by 1 unit of previous period
df_prev1 = Series_df.shift(12) # Look back 12 month
df_prev3 = Series_df.shift(12) # Look back 12 months
df_prev12 = Series_df.shift(12) # Look back 12 months
# Calculate rolling averages for previous year
Series_df[item+'1M_prev'] = df_prev1[item+'value'] # Take the value from 1 month ago
Series_df[item+'3MRoll_prev'] = df_prev3[item+'value'].rolling(window=3).mean() # Take the mean value of the last 3 months from 3 months ago
Series_df[item+'12MRoll_prev'] = df_prev12[item+'value'].rolling(window=12).mean() # Take the mean value of the last 12 months from 12 months ago
# Calculate percent change in rolling averages
Series_df[item+'1M_CHG_PERC'] = (Series_df[item+'value'] - Series_df[item+'1M_prev'] ) / Series_df[item+'1M_prev'] # 1 month % change relative to past 1 month
Series_df[item+'3MRoll_CHG_PERC'] = (Series_df[item+'3MRoll'] - Series_df[item+'3MRoll_prev']) / Series_df[item+'3MRoll_prev'] # 3 months % change relative to past 3 months
Series_df[item+'12MRoll_CHG_PERC'] = (Series_df[item+'12MRoll'] - Series_df[item+'12MRoll_prev']) / Series_df[item+'12MRoll_prev'] # 12 months % change relative to past 12 months
if runmode==1:
# Plot the time series
plt.figure(figsize=(12, 6))
plt.plot(Series_df['date'], Series_df[item+'1M_CHG_PERC'], label='1 Month % Change')
plt.plot(Series_df['date'], Series_df[item+'3MRoll_CHG_PERC'], label='3 Month Rolling Average % Change')
plt.plot(Series_df['date'], Series_df[item+'12MRoll_CHG_PERC'], label='12 Month Rolling Average % Change')
plt.xlabel('Date')
plt.ylabel('Percent Change')
plt.title('Percent Change in Rolling Averages'+Series_df[item+'Series'].iloc[1,])
plt.legend()
plt.grid(True)
plt.xticks(rotation=45)
plt.tight_layout()
plt.show()
# Store the dataframe in the dictionary
item_series_dfs[item + '_df'] = Series_df
return pd.DataFrame(Series_df)
Create an empty dataframe for the results from FRED
fredresults_df = pd.DataFrame()
Create a WIDE format result with FRED Series information, this also will become teh source for item_series_dfs which is a dictionary of dataframes (item_series_dfs)
fredresults_df = pd.concat([get_series(item) for item in dict_series_list], ignore_index=True)
RETURN a dict of dataFRAMES we can process and merge seperately
Sort the dictionary by row count in descending order
sorted_dfs = sorted(item_series_dfs.items(), key=lambda x: len(x[1]), reverse=True)
Initialize the merged dataframe with the first dataframe
merged_df = sorted_dfs[0][1]
Left join the remaining dataframes in sorted order with explicit suffixes
for i, (, df) in enumerate(sorted_dfs[1:], start=2):
suffix = f’{i}’
merged_df = pd.merge(merged_df, df, on=[‘Year’, ‘Month’], how=‘left’, suffixes=(‘’, suffix))
Drop duplicate rows based on ‘Year’ and ‘Month’, keeping the earliest ‘date’ value
merged_df = merged_df.sort_values(‘date’).drop_duplicates(subset=[‘Year’, ‘Month’], keep=‘first’)
Reset the index
merged_df.reset_index(drop=True, inplace=True)
Identify and rename duplicate columns in merged_df
merged_df = merged_df.loc[:,~merged_df.columns.duplicated()]
Identify and rename duplicate columns in merged_df
merged_df = merged_df.loc[:,~merged_df.columns.duplicated()]
Drop columns starting with ‘date’ and ‘value’
merged_df = merged_df.loc[:,~merged_df.columns.str.startswith(‘date’)]
fredresults_df = merged_df.loc[:,~merged_df.columns.str.startswith(‘value’)]
Display the final dataframe
if runmode==1:
print(fredresults_df)
Merge the Series data to the transaction data
Left join st_df to fredresults_df on ‘Year’ and ‘Month’
st_df = st_df.merge(fredresults_df, on=[‘Year’, ‘Month’], how=‘left’)
Display the result
print(st_df)
Push the table out to KNIME
if runmode==1:
print(fredresults_df.columns)
# Create a DataFrame with just the column headings and data types
column_info = pd.DataFrame({
‘Column’: fredresults_df.columns,
‘Data Type’: fredresults_df.dtypes
})
# Write the DataFrame to a CSV file
column_info.to_csv(output_path+'fredresults_column_info.csv', index=False)
st_df.to_csv(input_path+"InitialAssembly100_PCT_FREDSeriesInc.csv", index=False)
else:
knio.output_tables[0] = knio.Table.from_pandas(st_df)