Hi Everyone ![]()
I’m a bit late to post my solution for this one, but I have spent a few days implementing several models ![]()
Firstly, I would like to share the link to a paper that was written based on this data:
It’s a nice read and talks through training and applying both a Logistic Regression Model and a Decision Tree (DT).
Hence in my solution this week, I decided to use a DT to determine the variable importance when predicting Diabetes. Just to be clear and to not cause confusion with KNIME variables, I’m using the word variable(s) in the statistical sense to describe the columns in the table.
I have also tested 3 other models (Random Forest, XGBoost and Logistic Regression) to compare the predictive capabilities of the models firstly with only the most important variables and secondly with all variables.
The original data made of 768 samples and 9 columns, uses 0 to represent missing values, so I used the -Math Formula (Multi Column)- node to convert these to missing values in the workflow. In order to handle these missing values, I decided to remove two columns with large amounts of them: Insulin (374/768) and SkinThickness (227/768). After removal of these columns, I then removed any remaining rows that still contained missing values, a total of only 44 rows. The reason for removing the columns first, was to significantly reduce the number of rows containing missing values and to preserve as many rows from the dataset as possible.
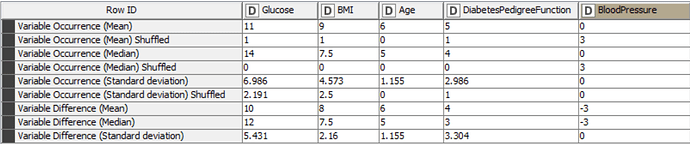
Before training any models, I have partitioned the data into internal and external sets. I have used K-Fold Cross validation on the internal set in order to run the DT model 5 times and gain Mean, Median and Standard Deviation values, using the -X-Partitioning- and -Loop End- nodes. Inside this loop, I have determined the Feature Importance by comparing the occurrence of each variable in the rule set of the decision tree against its shuffled counterpart. If the shuffled version appears more times in the DT rules than the non-shuffled version, then the variable difference becomes negative and it can be concluded that the variable is not important. The mean, median and standard deviation of variable occurrence and variable difference values can be viewed in the table below:
It is clear to see that the variable “Pregnancies” is not present in this table and this is due to the fact that it did not appear in any of the decision tree rule sets, neither in its original nor shuffled form. Therefore, it is possible to assume that this variable is not important in the prediction of diabetes based on this dataset. The variable “BloodPressure” appeared in rule sets as its shuffled form, but never in its original non-shuffled form, resulting in a negative variable difference. Again, this allows us to assume that this variable is not important in the model.
This leaves 4 variables, listed here in order of importance based on the variable occurrence; Glucose, BMI, Age and DiabetesPedigreeFunction. The most important being Glucose with the highest variable occurrence and variable difference. These important features also align with those determined by the DT model in the above scientific paper. However, it is important to say at this point, that further statistical analysis would be required to confirm these assumptions and any others made in the following paragraphs.
Following on from this, I implemented Random Forest, XGBoost and Logistic Regression models, firstly using only the 4 important variables and secondly using all the variables. I trained the models on the internal set using 5-fold cross validation and applied the models to the external set to predict the diabetes outcome.
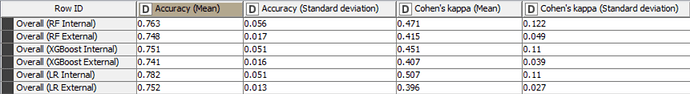
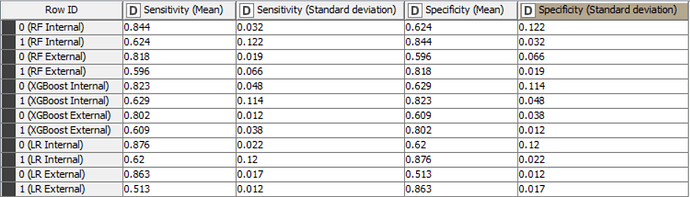
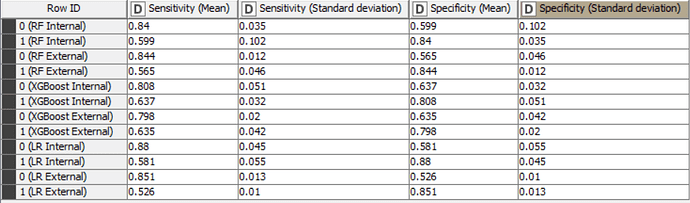
The Accuracy, Cohen’s Kappa, Specificity and Sensitivity results for internal and external sets are as follows for the models trained with only important variables:
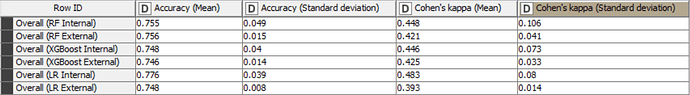
Here are the statistics for the models trained with all variables:
In fact for tree-based ensembles, the selection of important variables prior to training a model should not be necessary as they already have this capability built-in. For logistic regression models, it may be beneficial to create models with only important variables. However, in this case, the differences in statistical values don’t appear to be significant. Despite the non-significant difference in this example, it should be noted that it is useful to create models with only important variables, as this always improves the explicability of the model.
The accuracy of all models is inline with statistical results in the scientific paper and are higher than the baseline accuracy set by KNIME in this challenge.
This was a really interesting and enjoyable challenge ![]()
You can find my workflow here on the hub:
Enjoy it!
Heather