Hallo,
I updated to 4.7 the last few days and was very pleased with the geospatial extension. I’ve been waiting for this feature for years. There was a lot of curiosity and I immediately started trying out the tools. I was also impressed by the examples, congratulations.
I hope the text is understandable. Used Google translator German English
First attempt:
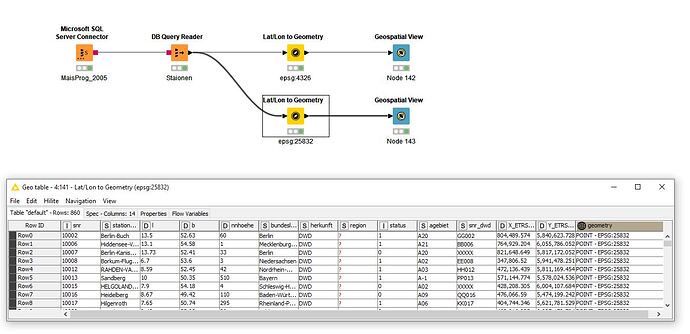
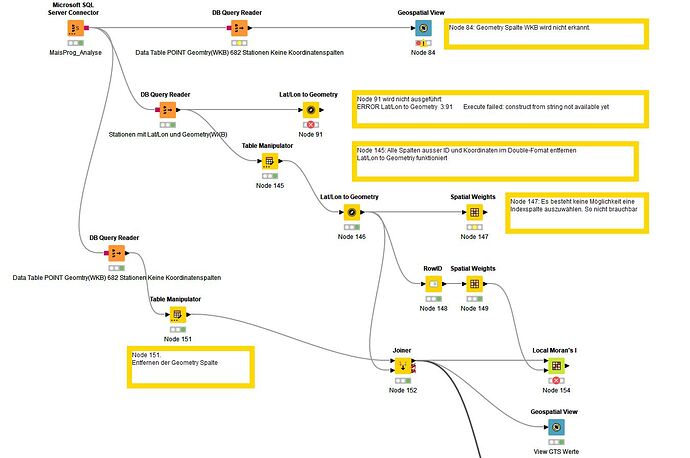
was quickly done Table from SQL Server Lat/Lon to geometry and display.
Table does not contain a geometry column.
Second try:
Test of the Spatial Data Analysis Tools, in particular Local Moran’s I Statistics and spatial weights matrix
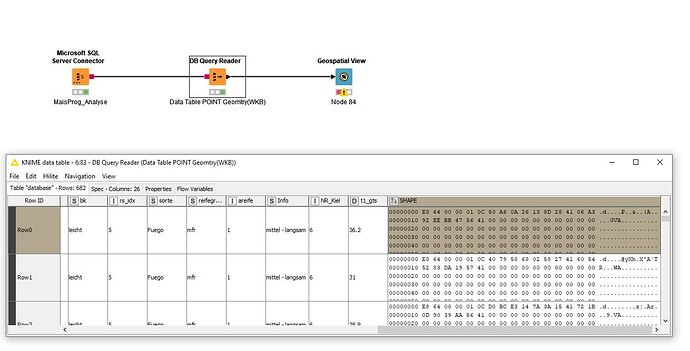
Tablefrom SQL Server with a geometry column, WKB format
Geospatail View faild.
Geometry column is not recognized
Third try:
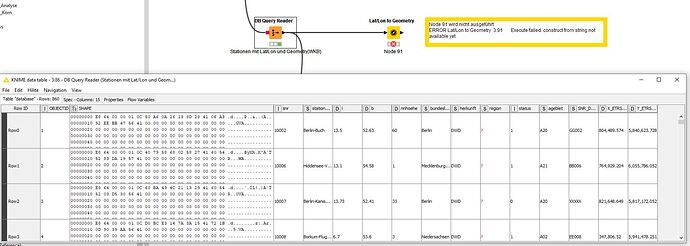
Load the station list from the DB. This contains the coordinates in WGS and UTM32N as well as a Geometry column.
A direct creation of the geometry fails.
WARN Lat/Lon to Geometry 3:91 Traceback (most recent call last):
File “C:\Program Files\KNIME\plugins\org.knime.python3.nodes_4.7.0.v202211291148\src\main\python_node_backend_launcher.py”, line 459, in execute
outputs = self._node.execute(exec_context, *inputs)
File “C:\Program Files\KNIME\plugins\sdl.harvard.geospatial_1.0.0.202212100353\src\main\python\src\nodes\conversion.py”, line 337, in execute
return knut.to_table(gdf, exec_context)
File “C:\Program Files\KNIME\plugins\sdl.harvard.geospatial_1.0.0.202212100353\src\main\python\src\util\knime_utils.py”, line 391, in to_table
return knext.Table.from_pandas(gdf)
File “C:\Program Files\KNIME\plugins\org.knime.python3_4.7.0.v202211291350\src\main\python\knime\api\table.py”, line 341, in from_pandas
return _backend.create_table_from_pandas(data, sentinel, row_ids=row_ids)
File “C:\Program Files\KNIME\plugins\org.knime.python3.arrow_4.7.0.v202211291117\src\main\python\knime_arrow_table.py”, line 136, in create_table_from_pandas
return _create_table_from_pandas(data, sentinel, row_ids)
File “C:\Program Files\KNIME\plugins\org.knime.python3.arrow_4.7.0.v202211291117\src\main\python\knime_arrow_table.py”, line 121, in _create_table_from_pandas
data = kap.pandas_df_to_arrow(data, row_ids=pandas_row_ids)
File “C:\Program Files\KNIME\plugins\org.knime.python3.arrow_4.7.0.v202211291117\src\main\python\knime_arrow_pandas.py”, line 77, in pandas_df_to_arrow
df = convert_df_to_ktypes_from_schema(data_frame, schema)
File “C:\Program Files\KNIME\plugins\org.knime.python3.arrow_4.7.0.v202211291117\src\main\python\knime_arrow_pandas.py”, line 178, in convert_df_to_ktypes_from_schema
df = df.copy(deep=False)
File “C:\Program Files\KNIME\plugins\sdl.harvard.geospatial.channel.bin.win32.x86_64_1.0.0.202212100353\env\lib\site-packages\pandas\core\generic.py”, line 6370, in copy
return self._constructor(data).finalize(self, method=“copy”)
File “C:\Program Files\KNIME\plugins\sdl.harvard.geospatial.channel.bin.win32.x86_64_1.0.0.202212100353\env\lib\site-packages\geopandas\geodataframe.py”, line 33, in _geodataframe_constructor_with_fallback
geometry_cols_mask = df.dtypes == “geometry”
File “C:\Program Files\KNIME\plugins\sdl.harvard.geospatial.channel.bin.win32.x86_64_1.0.0.202212100353\env\lib\site-packages\pandas\core\ops\common.py”, line 72, in new_method
return method(self, other)
File “C:\Program Files\KNIME\plugins\sdl.harvard.geospatial.channel.bin.win32.x86_64_1.0.0.202212100353\env\lib\site-packages\pandas\core\arraylike.py”, line 42, in eq
return self._cmp_method(other, operator.eq)
File “C:\Program Files\KNIME\plugins\sdl.harvard.geospatial.channel.bin.win32.x86_64_1.0.0.202212100353\env\lib\site-packages\pandas\core\series.py”, line 6243, in _cmp_method
res_values = ops.comparison_op(lvalues, rvalues, op)
File “C:\Program Files\KNIME\plugins\sdl.harvard.geospatial.channel.bin.win32.x86_64_1.0.0.202212100353\env\lib\site-packages\pandas\core\ops\array_ops.py”, line 287, in comparison_op
res_values = comp_method_OBJECT_ARRAY(op, lvalues, rvalues)
File “C:\Program Files\KNIME\plugins\sdl.harvard.geospatial.channel.bin.win32.x86_64_1.0.0.202212100353\env\lib\site-packages\pandas\core\ops\array_ops.py”, line 75, in comp_method_OBJECT_ARRAY
result = libops.scalar_compare(x.ravel(), y, op)
File “pandas_libs\ops.pyx”, line 95, in pandas._libs.ops.scalar_compare
File “C:\Program Files\KNIME\plugins\sdl.harvard.geospatial.channel.bin.win32.x86_64_1.0.0.202212100353\env\lib\site-packages\pandas\core\dtypes\base.py”, line 126, in eq
other = self.construct_from_string(other)
File “C:\Program Files\KNIME\plugins\org.knime.python3.arrow_4.7.0.v202211291117\src\main\python\knime_arrow_pandas.py”, line 305, in construct_from_string
raise NotImplementedError(“construct from string not available yet”)
NotImplementedError: construct from string not available yet
ERROR Lat/Lon to Geometry 3:91 Execute failed: construct from string not available yet
Fourth attempt:
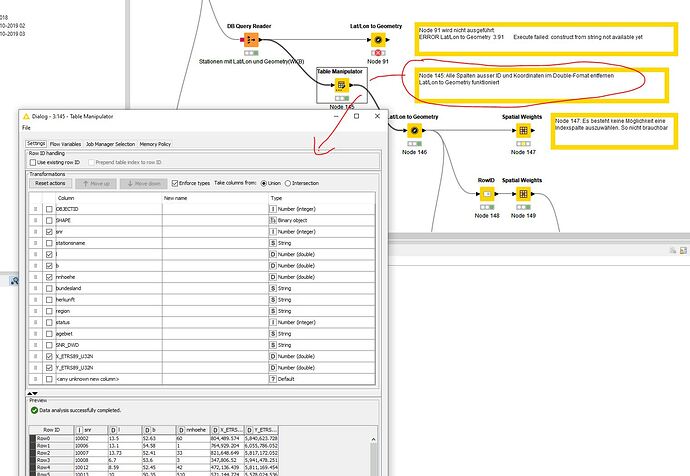
Table cleaned, unnecessary columns removed it works. Lat/Lon to Geoetry works.
Test spatial weights:
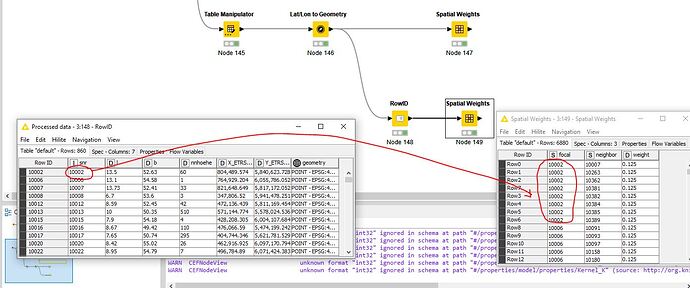
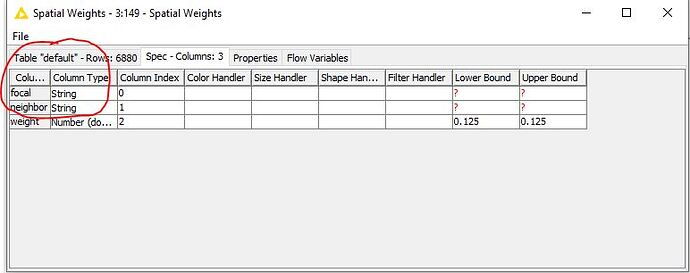
First attempt, what is noticeable, the selection of an ID column is missing. I can’t do anything with the row ID.
Can be avoided by assigning a column to the row ID. Node 148 and Node 149
For the evaluation, the assignment of the n neighbors must be comprehensible.
Fifth attempt:
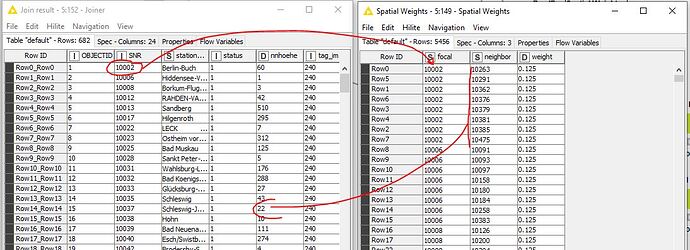
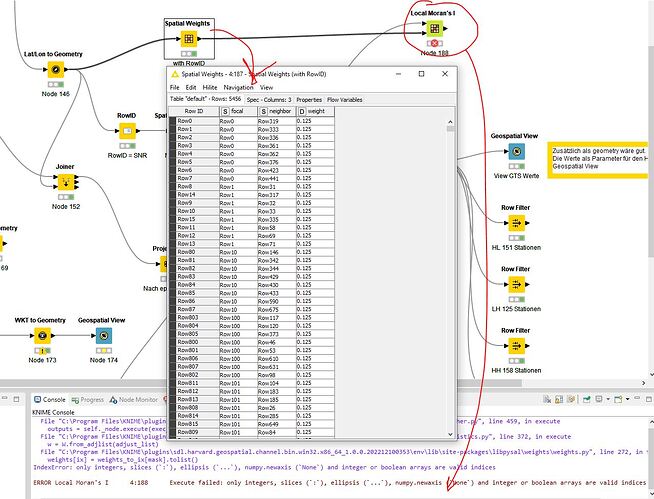
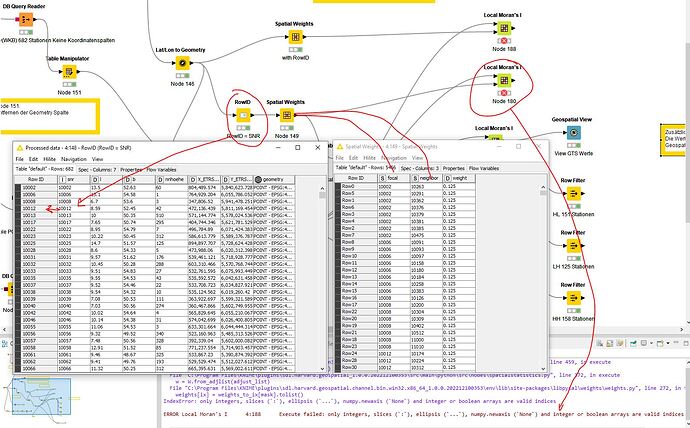
Prepared the data table and created spatial weights. What was immediately noticeable when configuring Local Moran’s I Statistics was that no relationship can be defined from the Spatial Weights table to the Data table. How is that supposed to work.
SNR (station number) from data table for focal and neighbor, which were derived from SNR, in the spatial weights table
Test Local Moran’s I
WARN Local Moran’s I 3:154 Traceback (most recent call last):
File “C:\Program Files\KNIME\plugins\org.knime.python3.nodes_4.7.0.v202211291148\src\main\python_node_backend_launcher.py”, line 459, in execute
outputs = self._node.execute(exec_context, *inputs)
File “C:\Program Files\KNIME\plugins\sdl.harvard.geospatial_1.0.0.202212100353\src\main\python\src\nodes\spatialstatistics.py”, line 372, in execute
w = W.from_adjlist(adjust_list)
File “C:\Program Files\KNIME\plugins\sdl.harvard.geospatial.channel.bin.win32.x86_64_1.0.0.202212100353\env\lib\site-packages\libpysal\weights\weights.py”, line 272, in from_adjlist
weights[ix] = weights_to_ix[mask].tolist()
IndexError: only integers, slices (:), ellipsis (...), numpy.newaxis (None) and integer or boolean arrays are valid indices
ERROR Local Moran’s I 3:154 Execute failed: only integers, slices (:), ellipsis (...), numpy.newaxis (None) and integer or boolean arrays are valid indices
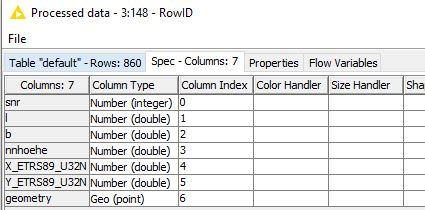
The table format for the spatial weights
Is the error here. The RowID was set from this data (SNR) in order to generate spatial weights.
Fazit:
Data is always in SQL Server 2019 and is prepared using procedures and views for further processing, including geometry.
OGC standards for geometry on columns are adopted to use them.
If a custom format is required for geomtry, then “WKB to Geomtry” and “Geometry to WKB” must also be added.
I could not test the writing back of the evaluated data in a GeoPacke. Would be enough, but means that the data then has to be transferred to the SQL server using another tool.
Here it would make sense to add DB Spatial Writer. This should then also be able to create a spatial index.
About spatial weights:
Here it must be possible to select an ID column in order to establish the relationship between spatial weights focal and the data table index (SNR) and neighbor (SNR). It is not clear how this relationship is established.
About the Geospatial Views:
A home button should be added over the extent (bondary) of the visualized data.
The representation should be able to be passed to the “Image to Report” node.
Local Moran’s I Statistics:
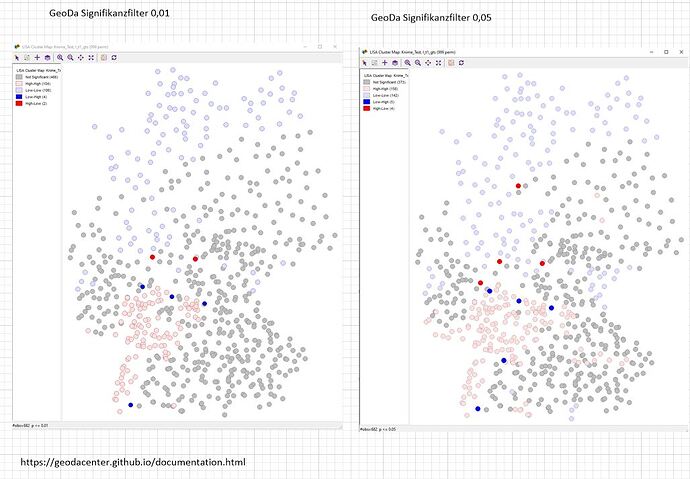
Geoda also offers permutation and significance settings. Should be available.
That’s it for now.
Patrik