Not exactly sure what you mean by installed by both R and RStudio. Typically you should only have one R installation with one library. You could try and check your version’s library with this comand:
.libPaths()
The error messages hint that you do not have the relevant package docxtractr installed.
And you will have to install RServe in the latest version. One thing I had success wit recently is described here:
After you have done that you could tell RStudio to find the Rtools necessary to compile new packages
devtools::find_rtools()
To repeat that step by step:
- install the latest stable version of Rtools to the directory c:\Rtools (no fancy path please) and Devtools
- set the environments of the path to RTools accoring to this entry
- set the PATH to RTools in the “.Renviron” file by typing:
usethis::edit_r_environ()
this should open the “.Renviron” file (yes with a dot at the beginning). There you should be able to enter the necessary PATH
PATH=“C:\Rtools\bin;${PATH}”
- run the selection of the path
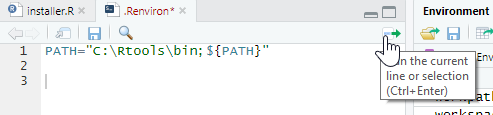
- tell RStudio to find RTools
devtools::find_rtools()
this should tell you that RTools is now active and you can use that. Now you can try to install RServe in the latest version again. Like this
install.packages(“Rserve”, “http://rforge.net/”, type = “source”, INSTALL_opts = “–no-multiarch”))
This:
install.packages(‘Rserve’,“http://rforge.net/",type="source”
Or this (after you downloaded the .tar file and put it in the path. You have to change the path accoring to your local machine):
install.packages(‘~/Downloads/Rserve_1.8-6.tar’, repos = NULL, type=“source”)